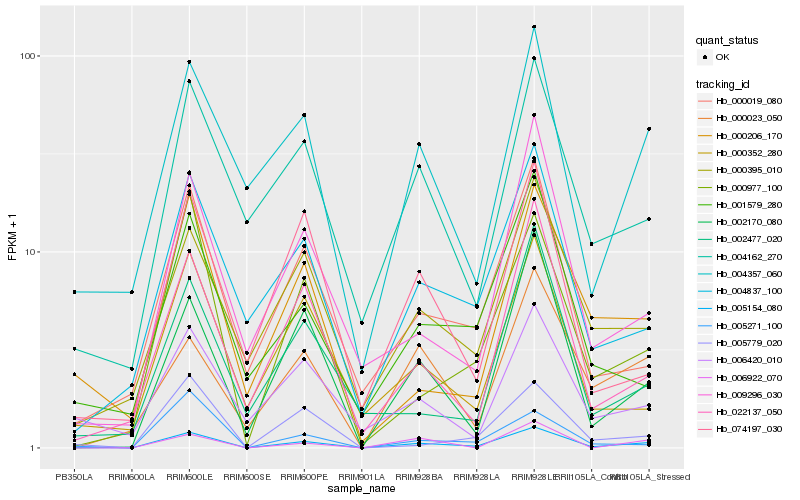
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005154_080 |
0.0 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105640764 [Jatropha curcas] |
| 2 |
Hb_002170_080 |
0.1776787703 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 3 |
Hb_022137_050 |
0.1961569082 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 1-like [Jatropha curcas] |
| 4 |
Hb_004357_060 |
0.1979585573 |
- |
- |
PREDICTED: uncharacterized protein At4g22758 [Jatropha curcas] |
| 5 |
Hb_000977_100 |
0.1990217453 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: acetolactate synthase small subunit 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_004837_100 |
0.2006031591 |
- |
- |
PREDICTED: dicarboxylate transporter 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000019_080 |
0.2041904128 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor ICE1 [Jatropha curcas] |
| 8 |
Hb_005779_020 |
0.2048153013 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004162_270 |
0.2052918017 |
- |
- |
PREDICTED: superoxide dismutase [Cu-Zn], chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_000395_010 |
0.2055293988 |
- |
- |
alpha/beta hydrolase domain containing protein 1,3, putative [Ricinus communis] |
| 11 |
Hb_009296_030 |
0.2095547342 |
- |
- |
PREDICTED: uncharacterized protein LOC105634328 [Jatropha curcas] |
| 12 |
Hb_000206_170 |
0.2100695771 |
- |
- |
hypothetical protein JCGZ_25514 [Jatropha curcas] |
| 13 |
Hb_000352_280 |
0.2133375372 |
- |
- |
PREDICTED: acetyl-coenzyme A synthetase, chloroplastic/glyoxysomal isoform X2 [Jatropha curcas] |
| 14 |
Hb_006922_070 |
0.2134248342 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 15 |
Hb_000023_050 |
0.215838761 |
- |
- |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 16 |
Hb_001579_280 |
0.2171504738 |
- |
- |
PREDICTED: 1,4-dihydroxy-2-naphthoyl-CoA synthase, peroxisomal-like [Jatropha curcas] |
| 17 |
Hb_006420_010 |
0.2187050451 |
- |
- |
PREDICTED: uncharacterized protein LOC105649815 [Jatropha curcas] |
| 18 |
Hb_074197_030 |
0.221500004 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_005271_100 |
0.2225766544 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_002477_020 |
0.2239970001 |
- |
- |
PREDICTED: uncharacterized protein LOC105631402 [Jatropha curcas] |