| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
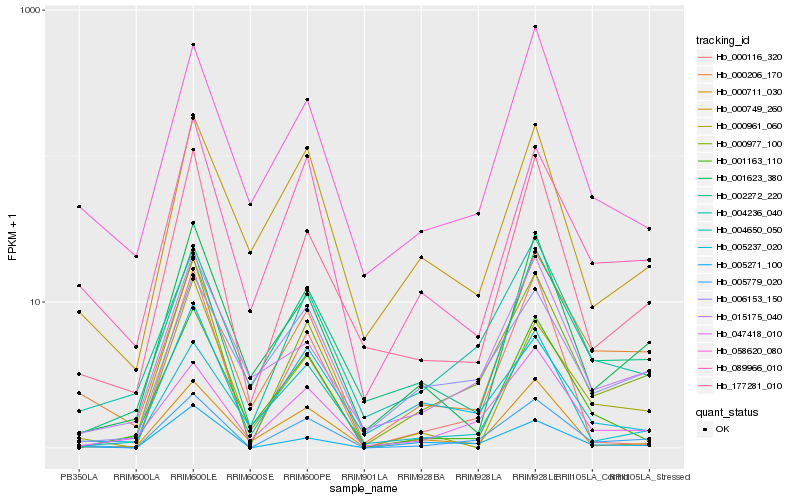
Hb_005779_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000977_100 |
0.1030737725 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: acetolactate synthase small subunit 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_004236_040 |
0.1591022268 |
- |
- |
biotin carboxyl carrier protein [Vernicia fordii] |
| 4 |
Hb_177281_010 |
0.159203809 |
- |
- |
PREDICTED: 50S ribosomal protein 5, chloroplastic-like [Citrus sinensis] |
| 5 |
Hb_047418_010 |
0.1625568115 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein P [Jatropha curcas] |
| 6 |
Hb_000206_170 |
0.1660677737 |
- |
- |
hypothetical protein JCGZ_25514 [Jatropha curcas] |
| 7 |
Hb_005271_100 |
0.1721798038 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_089966_010 |
0.1743519557 |
- |
- |
- |
| 9 |
Hb_000711_030 |
0.1851066512 |
- |
- |
hypothetical protein POPTR_0019s02860g [Populus trichocarpa] |
| 10 |
Hb_004650_050 |
0.1919361321 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_058620_080 |
0.1920881081 |
- |
- |
PREDICTED: phosphoglycerate kinase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001623_380 |
0.1932431354 |
- |
- |
PREDICTED: 7-hydroxymethyl chlorophyll a reductase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002272_220 |
0.1946155096 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005237_020 |
0.1951415388 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 15 |
Hb_000749_260 |
0.1952364599 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000116_320 |
0.1968367635 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 17 |
Hb_006153_150 |
0.1980623257 |
- |
- |
PREDICTED: uncharacterized protein LOC105645999 [Jatropha curcas] |
| 18 |
Hb_015175_040 |
0.200882102 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001163_110 |
0.2013723949 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PKL2 [Jatropha curcas] |
| 20 |
Hb_000961_060 |
0.2022406324 |
- |
- |
uvb-resistance protein uvr8, putative [Ricinus communis] |