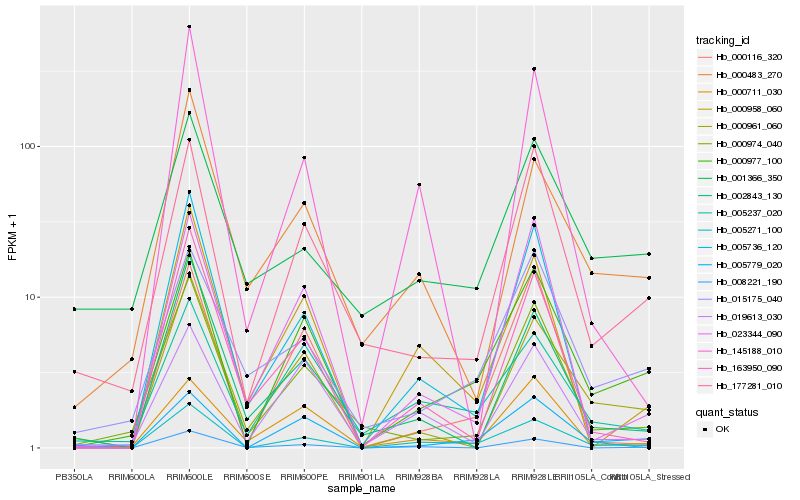
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005271_100 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_000977_100 |
0.1526736372 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: acetolactate synthase small subunit 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_005779_020 |
0.1721798038 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005237_020 |
0.1754579767 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 5 |
Hb_000958_060 |
0.1802089385 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_177281_010 |
0.1908249581 |
- |
- |
PREDICTED: 50S ribosomal protein 5, chloroplastic-like [Citrus sinensis] |
| 7 |
Hb_000961_060 |
0.19871116 |
- |
- |
uvb-resistance protein uvr8, putative [Ricinus communis] |
| 8 |
Hb_145188_010 |
0.2019034151 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 9 |
Hb_019613_030 |
0.2042800947 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase CLF [Jatropha curcas] |
| 10 |
Hb_000974_040 |
0.208644378 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g49770 [Jatropha curcas] |
| 11 |
Hb_002843_130 |
0.2087177869 |
- |
- |
PREDICTED: GRF1-interacting factor 2-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_005736_120 |
0.2092536106 |
- |
- |
PREDICTED: carboxyvinyl-carboxyphosphonate phosphorylmutase, chloroplastic [Gossypium raimondii] |
| 13 |
Hb_008221_190 |
0.2096388642 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 [Jatropha curcas] |
| 14 |
Hb_163950_090 |
0.2112580922 |
- |
- |
PREDICTED: traB domain-containing protein isoform X1 [Jatropha curcas] |
| 15 |
Hb_000483_270 |
0.2115834373 |
- |
- |
PREDICTED: uncharacterized protein LOC105632352 [Jatropha curcas] |
| 16 |
Hb_001366_350 |
0.2158032813 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase isoform X2 [Jatropha curcas] |
| 17 |
Hb_015175_040 |
0.2163653536 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000116_320 |
0.2168530841 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 19 |
Hb_000711_030 |
0.2180479133 |
- |
- |
hypothetical protein POPTR_0019s02860g [Populus trichocarpa] |
| 20 |
Hb_023344_090 |
0.2182551534 |
- |
- |
Photosystem II reaction center W protein, putative [Ricinus communis] |