| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
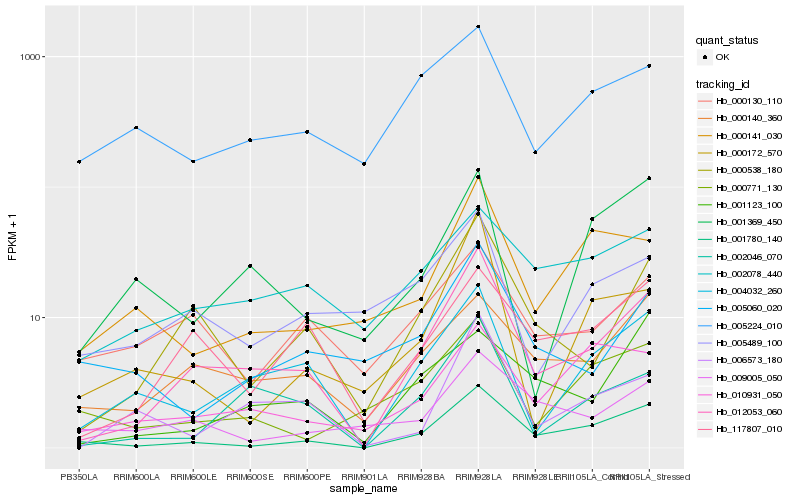
Hb_012053_060 |
0.0 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g44820 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000538_180 |
0.1532859183 |
- |
- |
PREDICTED: probable polyamine oxidase 4 [Jatropha curcas] |
| 3 |
Hb_001780_140 |
0.1722949798 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 4 |
Hb_002046_070 |
0.1734146153 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-induced protein 22D-like isoform X1 [Populus euphratica] |
| 5 |
Hb_004032_260 |
0.1841721362 |
- |
- |
PREDICTED: uncharacterized protein LOC105635147 [Jatropha curcas] |
| 6 |
Hb_117807_010 |
0.2070923561 |
- |
- |
PREDICTED: uncharacterized protein LOC105635720 [Jatropha curcas] |
| 7 |
Hb_000140_360 |
0.2073245196 |
- |
- |
PREDICTED: uncharacterized protein LOC105642906 [Jatropha curcas] |
| 8 |
Hb_006573_180 |
0.2149995199 |
transcription factor |
TF Family: WRKY |
hypothetical protein RCOM_1581940 [Ricinus communis] |
| 9 |
Hb_002078_440 |
0.2389348563 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005489_100 |
0.240820806 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000172_570 |
0.2433567771 |
- |
- |
PREDICTED: uncharacterized protein LOC105636518 isoform X1 [Jatropha curcas] |
| 12 |
Hb_010931_050 |
0.2439092495 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25-like [Jatropha curcas] |
| 13 |
Hb_001123_100 |
0.2441157815 |
- |
- |
PREDICTED: uncharacterized protein LOC105647337 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000141_030 |
0.2465454756 |
- |
- |
PREDICTED: scarecrow-like protein 22 [Jatropha curcas] |
| 15 |
Hb_000130_110 |
0.2479537958 |
- |
- |
PREDICTED: TPR repeat-containing thioredoxin TTL1 [Jatropha curcas] |
| 16 |
Hb_001369_450 |
0.2518911065 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 17 |
Hb_000771_130 |
0.2522160562 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 18 |
Hb_005060_020 |
0.2527225788 |
- |
- |
PREDICTED: metal tolerance protein 10-like isoform X1 [Populus euphratica] |
| 19 |
Hb_009005_050 |
0.2533228891 |
- |
- |
neutral/alkaline invertase 3 [Hevea brasiliensis] |
| 20 |
Hb_005224_010 |
0.253851683 |
- |
- |
glycine-rich protein [Bruguiera gymnorhiza] |