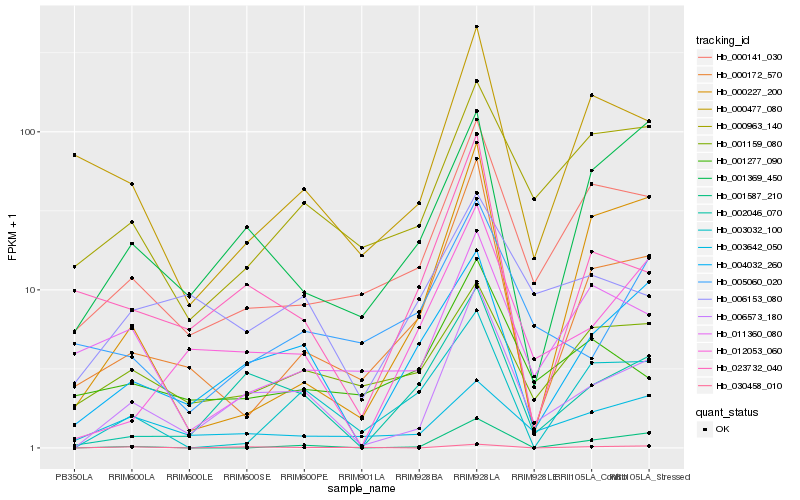
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006573_180 |
0.0 |
transcription factor |
TF Family: WRKY |
hypothetical protein RCOM_1581940 [Ricinus communis] |
| 2 |
Hb_002046_070 |
0.1850917801 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-induced protein 22D-like isoform X1 [Populus euphratica] |
| 3 |
Hb_004032_260 |
0.2121667841 |
- |
- |
PREDICTED: uncharacterized protein LOC105635147 [Jatropha curcas] |
| 4 |
Hb_012053_060 |
0.2149995199 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g44820 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000141_030 |
0.2377880815 |
- |
- |
PREDICTED: scarecrow-like protein 22 [Jatropha curcas] |
| 6 |
Hb_000172_570 |
0.243830565 |
- |
- |
PREDICTED: uncharacterized protein LOC105636518 isoform X1 [Jatropha curcas] |
| 7 |
Hb_023732_040 |
0.2453392507 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 8 |
Hb_000963_140 |
0.2583077513 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 9 |
Hb_000477_080 |
0.2651136612 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11 [Jatropha curcas] |
| 10 |
Hb_001369_450 |
0.2696898001 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 11 |
Hb_001587_210 |
0.271068024 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 [Jatropha curcas] |
| 12 |
Hb_006153_080 |
0.2739252519 |
- |
- |
flavin-containing monooxygenase-related family protein [Populus trichocarpa] |
| 13 |
Hb_001159_080 |
0.2753770911 |
- |
- |
hypothetical protein JCGZ_20745 [Jatropha curcas] |
| 14 |
Hb_001277_090 |
0.2761037649 |
- |
- |
PREDICTED: uncharacterized protein LOC105632061 [Jatropha curcas] |
| 15 |
Hb_003642_050 |
0.2798945347 |
- |
- |
hypothetical protein CISIN_1g048193mg, partial [Citrus sinensis] |
| 16 |
Hb_005060_020 |
0.2802066473 |
- |
- |
PREDICTED: metal tolerance protein 10-like isoform X1 [Populus euphratica] |
| 17 |
Hb_030458_010 |
0.2803913667 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 18 |
Hb_011360_080 |
0.2838074924 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_003032_100 |
0.2838431819 |
- |
- |
PREDICTED: GDP-mannose transporter GONST4 [Jatropha curcas] |
| 20 |
Hb_000227_200 |
0.2839177142 |
- |
- |
PREDICTED: protein FANTASTIC FOUR 3 [Populus euphratica] |