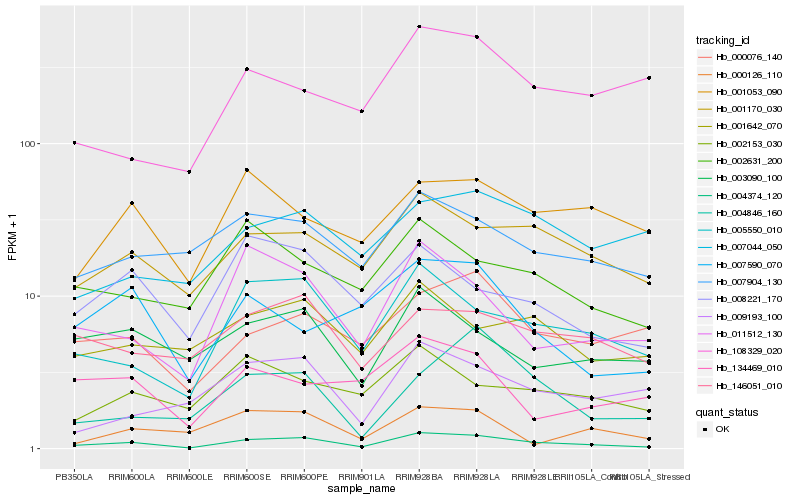
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004374_120 |
0.0 |
- |
- |
hypothetical protein JCGZ_20558 [Jatropha curcas] |
| 2 |
Hb_005550_010 |
0.1659936549 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 4-like [Jatropha curcas] |
| 3 |
Hb_001170_030 |
0.1810473109 |
- |
- |
synaptonemal complex protein, putative [Ricinus communis] |
| 4 |
Hb_011512_130 |
0.185364475 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Jatropha curcas] |
| 5 |
Hb_007904_130 |
0.2030381027 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 6 |
Hb_000076_140 |
0.2032579131 |
- |
- |
PREDICTED: glucose-6-phosphate/phosphate translocator 1, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_003090_100 |
0.2034822426 |
- |
- |
PREDICTED: uncharacterized protein LOC105646670 [Jatropha curcas] |
| 8 |
Hb_008221_170 |
0.2058108298 |
transcription factor |
TF Family: BES1 |
PREDICTED: BES1/BZR1 homolog protein 4 isoform X1 [Populus euphratica] |
| 9 |
Hb_146051_010 |
0.2063299047 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 10 |
Hb_002631_200 |
0.2078511496 |
- |
- |
PREDICTED: uncharacterized protein LOC105646171 isoform X2 [Jatropha curcas] |
| 11 |
Hb_001642_070 |
0.2082476056 |
- |
- |
PREDICTED: uncharacterized protein LOC105646431 [Jatropha curcas] |
| 12 |
Hb_009193_100 |
0.2091499956 |
- |
- |
hypothetical protein JCGZ_15709 [Jatropha curcas] |
| 13 |
Hb_002153_030 |
0.2120973304 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 14 |
Hb_001053_090 |
0.2127529862 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 30-like [Jatropha curcas] |
| 15 |
Hb_004846_160 |
0.2136953366 |
- |
- |
- |
| 16 |
Hb_007590_070 |
0.2142354341 |
- |
- |
PREDICTED: inositol 3-kinase [Jatropha curcas] |
| 17 |
Hb_000126_110 |
0.22223895 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X3 [Populus euphratica] |
| 18 |
Hb_108329_020 |
0.223889402 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 19 |
Hb_134469_010 |
0.224473364 |
- |
- |
PREDICTED: protein argonaute 5 isoform X1 [Jatropha curcas] |
| 20 |
Hb_007044_050 |
0.2255354176 |
- |
- |
PREDICTED: putative endo-1,3(4)-beta-glucanase 2 [Jatropha curcas] |