| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
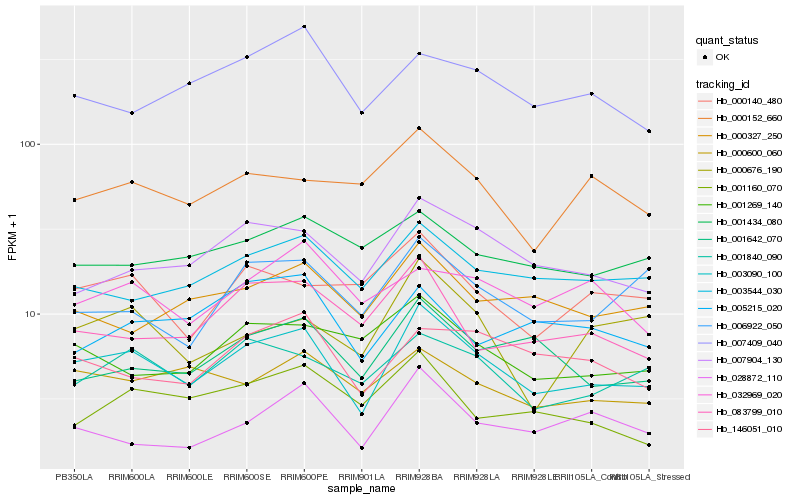
Hb_003090_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646670 [Jatropha curcas] |
| 2 |
Hb_007904_130 |
0.1222330144 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 3 |
Hb_001840_090 |
0.1268161944 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 4 |
Hb_003544_030 |
0.1309213793 |
- |
- |
PREDICTED: citrate synthase, glyoxysomal [Jatropha curcas] |
| 5 |
Hb_001642_070 |
0.1328762574 |
- |
- |
PREDICTED: uncharacterized protein LOC105646431 [Jatropha curcas] |
| 6 |
Hb_083799_010 |
0.1370120865 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 7 |
Hb_000600_060 |
0.1378287549 |
- |
- |
PREDICTED: uncharacterized protein LOC105633515 isoform X1 [Jatropha curcas] |
| 8 |
Hb_006922_050 |
0.1385147932 |
- |
- |
PREDICTED: dipeptidyl peptidase 8 [Jatropha curcas] |
| 9 |
Hb_146051_010 |
0.1387512344 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 10 |
Hb_000327_250 |
0.1396721299 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 11 |
Hb_001269_140 |
0.1409763922 |
- |
- |
PREDICTED: nuclear pore complex protein NUP96 [Jatropha curcas] |
| 12 |
Hb_028872_110 |
0.1429385733 |
- |
- |
PREDICTED: uncharacterized protein LOC105644776 [Jatropha curcas] |
| 13 |
Hb_000140_480 |
0.1436582733 |
- |
- |
PREDICTED: uncharacterized protein LOC105637350 [Jatropha curcas] |
| 14 |
Hb_000152_660 |
0.1443122059 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 7 [Jatropha curcas] |
| 15 |
Hb_000676_190 |
0.1446780102 |
- |
- |
hypothetical protein JCGZ_08279 [Jatropha curcas] |
| 16 |
Hb_007409_040 |
0.1448226644 |
- |
- |
actin family protein [Populus trichocarpa] |
| 17 |
Hb_001160_070 |
0.1471168143 |
- |
- |
PREDICTED: uncharacterized protein LOC105632163 [Jatropha curcas] |
| 18 |
Hb_001434_080 |
0.1477144526 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Jatropha curcas] |
| 19 |
Hb_005215_020 |
0.1508031364 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB44 [Jatropha curcas] |
| 20 |
Hb_032969_020 |
0.1516693281 |
- |
- |
conserved hypothetical protein [Ricinus communis] |