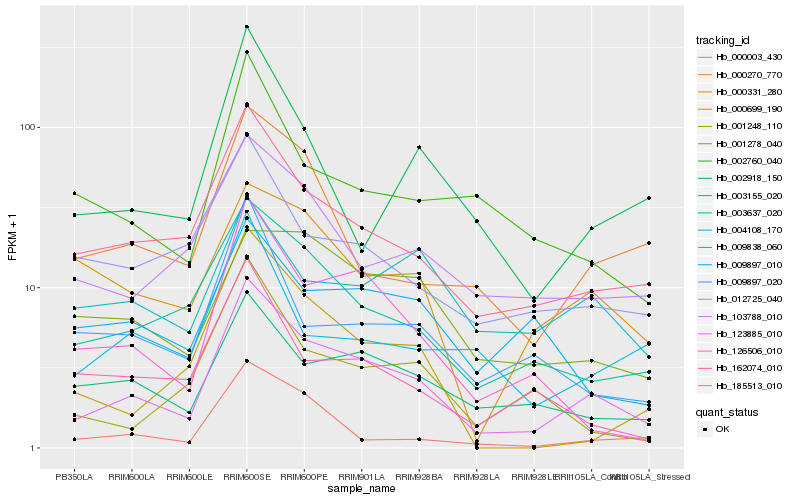
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_123885_010 |
0.0 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 2 |
Hb_185513_010 |
0.1565520573 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003637_020 |
0.1707100977 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003155_020 |
0.1838186435 |
- |
- |
Rpp4C3 [Medicago truncatula] |
| 5 |
Hb_126506_010 |
0.1870143199 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 6 |
Hb_103788_010 |
0.1935305173 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 7 |
Hb_000270_770 |
0.2049833561 |
- |
- |
PREDICTED: uncharacterized protein LOC105640410 [Jatropha curcas] |
| 8 |
Hb_012725_040 |
0.2057829866 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 9 |
Hb_002760_040 |
0.2087285694 |
- |
- |
PREDICTED: uncharacterized protein LOC105641234 [Jatropha curcas] |
| 10 |
Hb_009897_020 |
0.2097065338 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 11 |
Hb_002918_150 |
0.2116364405 |
transcription factor |
TF Family: ERF |
ethylene-responsive element binding protein 1 [Hevea brasiliensis] |
| 12 |
Hb_000003_430 |
0.2119268287 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 13 |
Hb_000331_280 |
0.2142209352 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 14 |
Hb_001248_110 |
0.2169054401 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 15 |
Hb_162074_010 |
0.2169287172 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 16 |
Hb_000699_190 |
0.2180432319 |
- |
- |
unknown [Populus trichocarpa] |
| 17 |
Hb_009838_060 |
0.2198525264 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004108_170 |
0.2209257865 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_009897_010 |
0.2235473414 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 20 |
Hb_001278_040 |
0.2246102176 |
- |
- |
PREDICTED: lipid phosphate phosphatase 1-like isoform X1 [Jatropha curcas] |