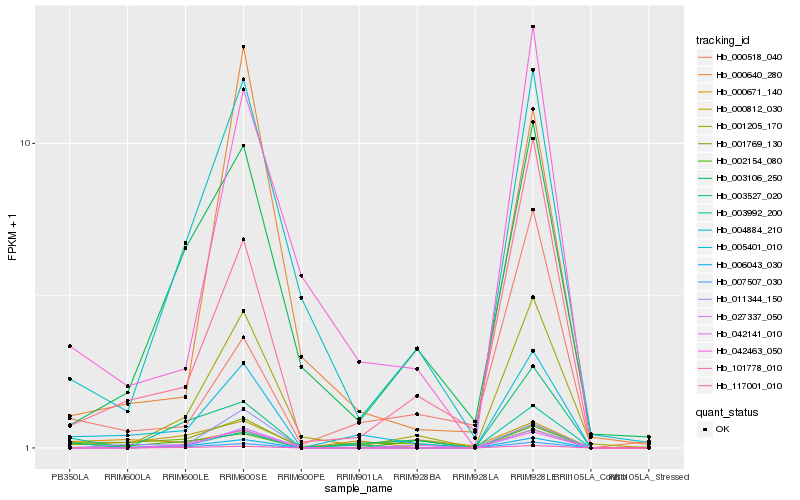
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005401_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_006043_030 |
0.1536890053 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 3 |
Hb_001769_130 |
0.2104233081 |
- |
- |
- |
| 4 |
Hb_117001_010 |
0.2137239078 |
- |
- |
- |
| 5 |
Hb_003527_020 |
0.2215048793 |
- |
- |
BnaC01g27150D [Brassica napus] |
| 6 |
Hb_000671_140 |
0.2216872377 |
- |
- |
hypothetical protein VITISV_003767 [Vitis vinifera] |
| 7 |
Hb_042463_050 |
0.2243540327 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004884_210 |
0.2389387929 |
- |
- |
PREDICTED: mitoferrin-like [Jatropha curcas] |
| 9 |
Hb_003106_250 |
0.2479506922 |
- |
- |
hypothetical protein POPTR_0008s13520g [Populus trichocarpa] |
| 10 |
Hb_001205_170 |
0.2493218986 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |
| 11 |
Hb_027337_050 |
0.2617451446 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 12 |
Hb_011344_150 |
0.2670400569 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 13 |
Hb_000812_030 |
0.2678323921 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 14 |
Hb_000640_280 |
0.2693043637 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 15 |
Hb_003992_200 |
0.2704227652 |
- |
- |
- |
| 16 |
Hb_007507_030 |
0.2704564809 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 17 |
Hb_000518_040 |
0.2709352238 |
- |
- |
- |
| 18 |
Hb_042141_010 |
0.274247223 |
- |
- |
- |
| 19 |
Hb_101778_010 |
0.2749353431 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 20 |
Hb_002154_080 |
0.2813699543 |
- |
- |
hypothetical protein MIMGU_mgv1a018017mg, partial [Erythranthe guttata] |