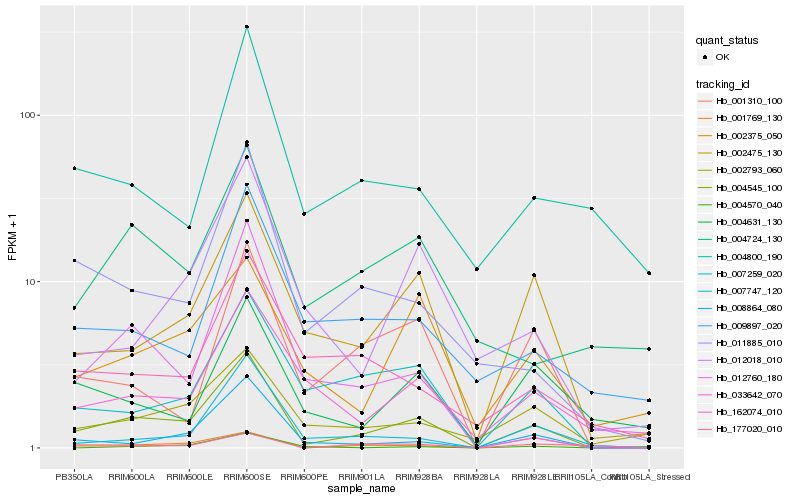
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004545_100 |
0.0 |
- |
- |
PREDICTED: protein BONZAI 3 [Jatropha curcas] |
| 2 |
Hb_002475_130 |
0.1955616854 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 4 [Jatropha curcas] |
| 3 |
Hb_008864_080 |
0.2101622719 |
- |
- |
PREDICTED: uncharacterized protein LOC105631804 [Jatropha curcas] |
| 4 |
Hb_007259_020 |
0.2164097607 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 [Jatropha curcas] |
| 5 |
Hb_002793_060 |
0.2215741523 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g02980 [Populus euphratica] |
| 6 |
Hb_011885_010 |
0.2265715382 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 7 |
Hb_033642_070 |
0.228171855 |
- |
- |
PREDICTED: oligopeptide transporter 7-like [Jatropha curcas] |
| 8 |
Hb_012760_180 |
0.2301873911 |
- |
- |
Basic-leucine zipper transcription factor family protein, putative [Theobroma cacao] |
| 9 |
Hb_007747_120 |
0.2348817004 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_012018_010 |
0.2383859505 |
- |
- |
hypothetical protein MIMGU_mgv1a0024251mg, partial [Erythranthe guttata] |
| 11 |
Hb_001310_100 |
0.2398261466 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable N-acetyltransferase HLS1 [Jatropha curcas] |
| 12 |
Hb_002375_050 |
0.2401505301 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 13 |
Hb_004724_130 |
0.2430599111 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14 [Jatropha curcas] |
| 14 |
Hb_009897_020 |
0.2437026609 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 15 |
Hb_001769_130 |
0.2458241461 |
- |
- |
- |
| 16 |
Hb_177020_010 |
0.2504227944 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |
| 17 |
Hb_004800_190 |
0.2510443143 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 4 [Jatropha curcas] |
| 18 |
Hb_162074_010 |
0.2520616226 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 19 |
Hb_004570_040 |
0.2521153372 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_004631_130 |
0.252134309 |
- |
- |
helicase domain-containing family protein [Populus trichocarpa] |