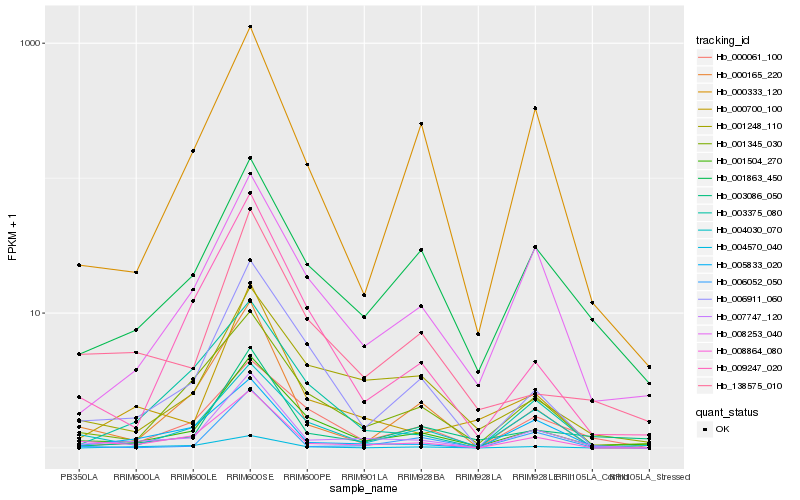
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007747_120 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_008864_080 |
0.1390408587 |
- |
- |
PREDICTED: uncharacterized protein LOC105631804 [Jatropha curcas] |
| 3 |
Hb_005833_020 |
0.163241156 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 4 |
Hb_000700_100 |
0.1748468548 |
- |
- |
PREDICTED: protein SULFUR DEFICIENCY-INDUCED 1 [Jatropha curcas] |
| 5 |
Hb_001504_270 |
0.1811824868 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 6 |
Hb_008253_040 |
0.1953843711 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 7 |
Hb_000165_220 |
0.1957717615 |
- |
- |
PREDICTED: taxadien-5-alpha-ol O-acetyltransferase [Jatropha curcas] |
| 8 |
Hb_006052_050 |
0.2020984757 |
- |
- |
hypothetical protein EUGRSUZ_A00568 [Eucalyptus grandis] |
| 9 |
Hb_001863_450 |
0.2055901876 |
- |
- |
cryptochrome 1.2 [Populus tremula] |
| 10 |
Hb_006911_060 |
0.2082227925 |
- |
- |
hypothetical protein CISIN_1g000594mg [Citrus sinensis] |
| 11 |
Hb_001345_030 |
0.2103921221 |
- |
- |
hypothetical protein POPTR_0007s10390g, partial [Populus trichocarpa] |
| 12 |
Hb_000333_120 |
0.2120822186 |
- |
- |
PREDICTED: thiol protease aleurain-like [Jatropha curcas] |
| 13 |
Hb_003086_050 |
0.2173052398 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 14 |
Hb_004030_070 |
0.2194245938 |
- |
- |
PREDICTED: uncharacterized protein LOC105632434 [Jatropha curcas] |
| 15 |
Hb_004570_040 |
0.2200358043 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_009247_020 |
0.2233697086 |
- |
- |
hypothetical protein JCGZ_04985 [Jatropha curcas] |
| 17 |
Hb_003375_080 |
0.224384817 |
- |
- |
PREDICTED: uncharacterized protein LOC105643382 isoform X1 [Jatropha curcas] |
| 18 |
Hb_138575_010 |
0.2251165624 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 19 |
Hb_000061_100 |
0.2257042318 |
- |
- |
hypothetical protein CISIN_1g0053742mg, partial [Citrus sinensis] |
| 20 |
Hb_001248_110 |
0.2263119776 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |