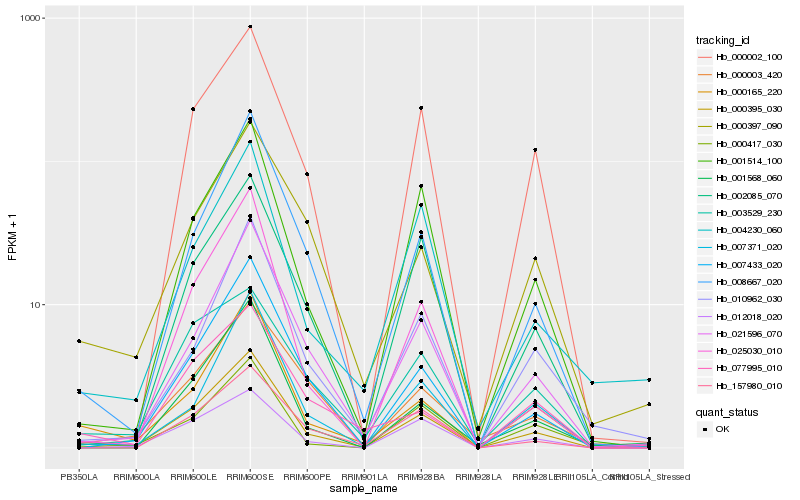
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000395_030 |
0.0 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 2 |
Hb_000165_220 |
0.1158818005 |
- |
- |
PREDICTED: taxadien-5-alpha-ol O-acetyltransferase [Jatropha curcas] |
| 3 |
Hb_021596_070 |
0.1256528279 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 4 |
Hb_008667_020 |
0.1275420813 |
- |
- |
PREDICTED: uncharacterized protein LOC101299609 [Fragaria vesca subsp. vesca] |
| 5 |
Hb_007433_020 |
0.1303474275 |
- |
- |
hypothetical protein VITISV_005320 [Vitis vinifera] |
| 6 |
Hb_000002_100 |
0.1367345108 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 7 |
Hb_001514_100 |
0.1370437669 |
- |
- |
PREDICTED: F-box protein GID2 [Jatropha curcas] |
| 8 |
Hb_002085_070 |
0.1451252351 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_000397_090 |
0.1462373051 |
- |
- |
PREDICTED: uncharacterized protein LOC105641235 [Jatropha curcas] |
| 10 |
Hb_001568_060 |
0.1485561412 |
- |
- |
PREDICTED: uncharacterized protein LOC105650728 [Jatropha curcas] |
| 11 |
Hb_007371_020 |
0.1489182539 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_012018_020 |
0.1510102479 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Eucalyptus grandis] |
| 13 |
Hb_010962_030 |
0.1513830607 |
- |
- |
kinase, putative [Ricinus communis] |
| 14 |
Hb_025030_010 |
0.1542971272 |
- |
- |
- |
| 15 |
Hb_004230_060 |
0.1555850007 |
- |
- |
PREDICTED: ferritin, chloroplastic-like [Populus euphratica] |
| 16 |
Hb_000417_030 |
0.1594156119 |
transcription factor |
TF Family: G2-like |
PREDICTED: putative two-component response regulator ARR19 [Jatropha curcas] |
| 17 |
Hb_003529_230 |
0.1608551792 |
transcription factor |
TF Family: ERF |
- |
| 18 |
Hb_000003_420 |
0.1613935387 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 19 |
Hb_157980_010 |
0.1620899128 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 20 |
Hb_077995_010 |
0.1634764638 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |