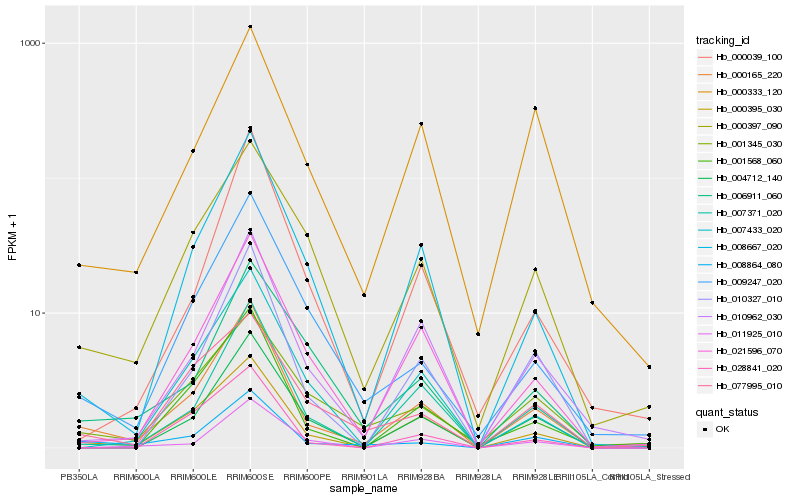
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000165_220 |
0.0 |
- |
- |
PREDICTED: taxadien-5-alpha-ol O-acetyltransferase [Jatropha curcas] |
| 2 |
Hb_000395_030 |
0.1158818005 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 3 |
Hb_008864_080 |
0.1221238763 |
- |
- |
PREDICTED: uncharacterized protein LOC105631804 [Jatropha curcas] |
| 4 |
Hb_007371_020 |
0.1317285244 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_008667_020 |
0.1417829893 |
- |
- |
PREDICTED: uncharacterized protein LOC101299609 [Fragaria vesca subsp. vesca] |
| 6 |
Hb_021596_070 |
0.1536753912 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 7 |
Hb_010962_030 |
0.1573245723 |
- |
- |
kinase, putative [Ricinus communis] |
| 8 |
Hb_010327_010 |
0.1605082667 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 9 |
Hb_004712_140 |
0.1660038717 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 10 |
Hb_000397_090 |
0.1670553758 |
- |
- |
PREDICTED: uncharacterized protein LOC105641235 [Jatropha curcas] |
| 11 |
Hb_009247_020 |
0.1677960468 |
- |
- |
hypothetical protein JCGZ_04985 [Jatropha curcas] |
| 12 |
Hb_007433_020 |
0.1695542886 |
- |
- |
hypothetical protein VITISV_005320 [Vitis vinifera] |
| 13 |
Hb_001568_060 |
0.1734303949 |
- |
- |
PREDICTED: uncharacterized protein LOC105650728 [Jatropha curcas] |
| 14 |
Hb_000333_120 |
0.1778040633 |
- |
- |
PREDICTED: thiol protease aleurain-like [Jatropha curcas] |
| 15 |
Hb_077995_010 |
0.1801885216 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 16 |
Hb_000039_100 |
0.1828372684 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 17 |
Hb_001345_030 |
0.1837048648 |
- |
- |
hypothetical protein POPTR_0007s10390g, partial [Populus trichocarpa] |
| 18 |
Hb_028841_020 |
0.1853109265 |
- |
- |
hypothetical protein CISIN_1g041345mg [Citrus sinensis] |
| 19 |
Hb_011925_010 |
0.185872289 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_006911_060 |
0.185938241 |
- |
- |
hypothetical protein CISIN_1g000594mg [Citrus sinensis] |