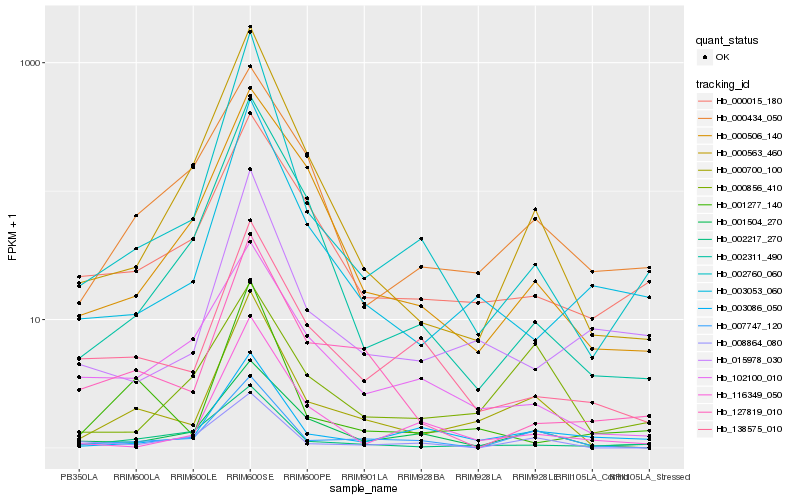
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000700_100 |
0.0 |
- |
- |
PREDICTED: protein SULFUR DEFICIENCY-INDUCED 1 [Jatropha curcas] |
| 2 |
Hb_007747_120 |
0.1748468548 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000563_460 |
0.204515157 |
- |
- |
PREDICTED: RING-H2 finger protein ATL2 [Jatropha curcas] |
| 4 |
Hb_001504_270 |
0.2085556262 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 5 |
Hb_002760_060 |
0.2156188184 |
- |
- |
PREDICTED: uncharacterized protein LOC105641235 [Jatropha curcas] |
| 6 |
Hb_003053_060 |
0.2204376836 |
- |
- |
PREDICTED: uncharacterized protein LOC105641515 [Jatropha curcas] |
| 7 |
Hb_003086_050 |
0.2260531907 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 8 |
Hb_138575_010 |
0.2278851147 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 9 |
Hb_000434_050 |
0.2287795601 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 10 |
Hb_102100_010 |
0.2290238225 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 11 |
Hb_001277_140 |
0.2292412681 |
- |
- |
PREDICTED: plasma membrane calcium-transporting ATPase 2-like [Takifugu rubripes] |
| 12 |
Hb_000506_140 |
0.2310020928 |
- |
- |
hypothetical protein PRUPE_ppb019425mg [Prunus persica] |
| 13 |
Hb_000015_180 |
0.2310713262 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 14 |
Hb_008864_080 |
0.2322426396 |
- |
- |
PREDICTED: uncharacterized protein LOC105631804 [Jatropha curcas] |
| 15 |
Hb_002311_490 |
0.2323241635 |
- |
- |
PREDICTED: RING-H2 finger protein ATL2 [Jatropha curcas] |
| 16 |
Hb_015978_030 |
0.2334370899 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 17 |
Hb_127819_010 |
0.2352555547 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 18 |
Hb_116349_050 |
0.2363523813 |
- |
- |
formin homology 2 domain-containing family protein [Populus trichocarpa] |
| 19 |
Hb_002217_270 |
0.2364229087 |
- |
- |
PREDICTED: desiccation-related protein PCC13-62-like [Jatropha curcas] |
| 20 |
Hb_000856_410 |
0.2370218538 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |