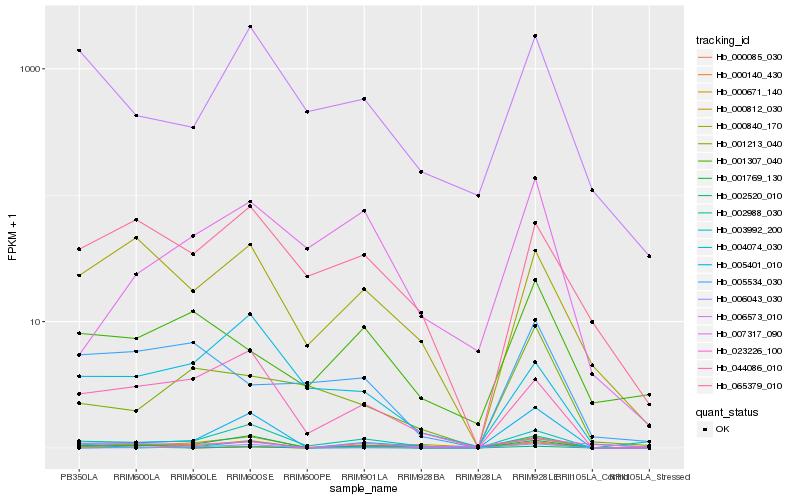
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000671_140 |
0.0 |
- |
- |
hypothetical protein VITISV_003767 [Vitis vinifera] |
| 2 |
Hb_005401_010 |
0.2216872377 |
- |
- |
- |
| 3 |
Hb_001769_130 |
0.2383252724 |
- |
- |
- |
| 4 |
Hb_044086_010 |
0.2500437783 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_000840_170 |
0.2787868411 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 6 |
Hb_006573_010 |
0.2980147461 |
- |
- |
heat shock protein [Hevea brasiliensis] |
| 7 |
Hb_000140_430 |
0.3007860922 |
- |
- |
- |
| 8 |
Hb_065379_010 |
0.3030546961 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 9 |
Hb_000085_030 |
0.3064962628 |
- |
- |
- |
| 10 |
Hb_002520_010 |
0.3082250222 |
- |
- |
PREDICTED: uncharacterized protein LOC105775392, partial [Gossypium raimondii] |
| 11 |
Hb_004074_030 |
0.3127242362 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_002988_030 |
0.3163798044 |
- |
- |
hypothetical protein EUGRSUZ_K02073 [Eucalyptus grandis] |
| 13 |
Hb_003992_200 |
0.3229268054 |
- |
- |
- |
| 14 |
Hb_006043_030 |
0.3249990517 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 15 |
Hb_000812_030 |
0.3280154804 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 16 |
Hb_023226_100 |
0.3288043087 |
- |
- |
PREDICTED: uncharacterized protein LOC104903172 [Beta vulgaris subsp. vulgaris] |
| 17 |
Hb_001307_040 |
0.3309287492 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 4B [Jatropha curcas] |
| 18 |
Hb_001213_040 |
0.3335966678 |
- |
- |
fructose-1,6-bisphosphatase, putative [Ricinus communis] |
| 19 |
Hb_007317_090 |
0.3350681999 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14 [Jatropha curcas] |
| 20 |
Hb_005534_030 |
0.335940827 |
- |
- |
hypothetical protein JCGZ_00732 [Jatropha curcas] |