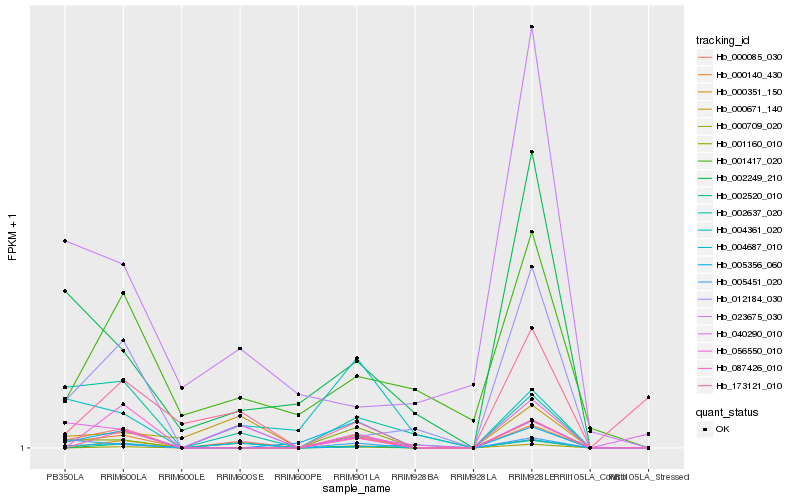
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000085_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_000140_430 |
0.0737580181 |
- |
- |
- |
| 3 |
Hb_012184_030 |
0.2553531817 |
- |
- |
- |
| 4 |
Hb_002637_020 |
0.2615124702 |
- |
- |
- |
| 5 |
Hb_002520_010 |
0.3054366822 |
- |
- |
PREDICTED: uncharacterized protein LOC105775392, partial [Gossypium raimondii] |
| 6 |
Hb_000671_140 |
0.3064962628 |
- |
- |
hypothetical protein VITISV_003767 [Vitis vinifera] |
| 7 |
Hb_001160_010 |
0.3085456378 |
- |
- |
- |
| 8 |
Hb_002249_210 |
0.3150309643 |
- |
- |
- |
| 9 |
Hb_005451_020 |
0.3203686583 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 10 |
Hb_004687_010 |
0.324545974 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 11 |
Hb_087426_010 |
0.3289123278 |
- |
- |
PREDICTED: uncharacterized protein LOC105803348 [Gossypium raimondii] |
| 12 |
Hb_001417_020 |
0.3358085896 |
- |
- |
PREDICTED: uncharacterized protein LOC104799692 [Tarenaya hassleriana] |
| 13 |
Hb_056550_010 |
0.3358430979 |
- |
- |
PREDICTED: uncharacterized protein LOC104112577 [Nicotiana tomentosiformis] |
| 14 |
Hb_000351_150 |
0.3384774697 |
- |
- |
Wall-associated kinase 2, putative [Theobroma cacao] |
| 15 |
Hb_005356_060 |
0.349149242 |
- |
- |
hypothetical protein POPTR_0013s14620g, partial [Populus trichocarpa] |
| 16 |
Hb_040290_010 |
0.3506982992 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 17 |
Hb_000709_020 |
0.3544774931 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 18 |
Hb_023675_030 |
0.3594701265 |
- |
- |
- |
| 19 |
Hb_173121_010 |
0.3597420347 |
- |
- |
- |
| 20 |
Hb_004361_020 |
0.3615747598 |
- |
- |
PREDICTED: uncharacterized protein LOC102625775 [Citrus sinensis] |