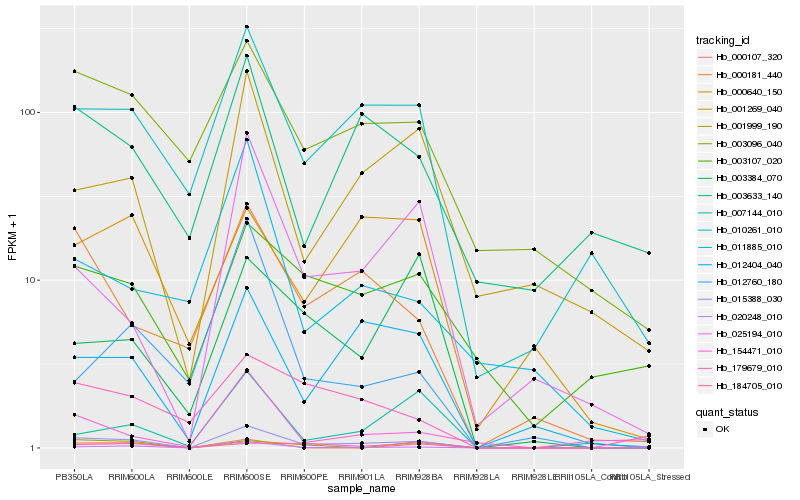
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015388_030 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_010261_010 |
0.2025795415 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |
| 3 |
Hb_012404_040 |
0.2202975249 |
- |
- |
PREDICTED: 1,4-alpha-glucan-branching enzyme 2-2, chloroplastic/amyloplastic [Jatropha curcas] |
| 4 |
Hb_000181_440 |
0.2355809114 |
- |
- |
- |
| 5 |
Hb_003096_040 |
0.2451384414 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0005s07290g [Populus trichocarpa] |
| 6 |
Hb_001269_040 |
0.2455179649 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 7 |
Hb_025194_010 |
0.2554133687 |
- |
- |
PREDICTED: uncharacterized protein LOC105644857 [Jatropha curcas] |
| 8 |
Hb_020248_010 |
0.258922828 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_179679_010 |
0.2666216943 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Fragaria vesca subsp. vesca] |
| 10 |
Hb_184705_010 |
0.2689355811 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_003633_140 |
0.2730464986 |
- |
- |
PREDICTED: uncharacterized protein LOC105647486 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003107_020 |
0.2781103168 |
- |
- |
hypothetical protein JCGZ_18656 [Jatropha curcas] |
| 13 |
Hb_154471_010 |
0.2802674959 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 14 |
Hb_007144_010 |
0.2806352246 |
- |
- |
- |
| 15 |
Hb_011885_010 |
0.2862970554 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 16 |
Hb_000107_320 |
0.2881369165 |
- |
- |
hypothetical protein POPTR_0004s21930g [Populus trichocarpa] |
| 17 |
Hb_000640_150 |
0.2884394833 |
transcription factor |
TF Family: WRKY |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001999_190 |
0.2897925551 |
- |
- |
GDP-mannose transporter, putative [Ricinus communis] |
| 19 |
Hb_003384_070 |
0.2905851977 |
- |
- |
PREDICTED: methylesterase 17-like [Populus euphratica] |
| 20 |
Hb_012760_180 |
0.2916499988 |
- |
- |
Basic-leucine zipper transcription factor family protein, putative [Theobroma cacao] |