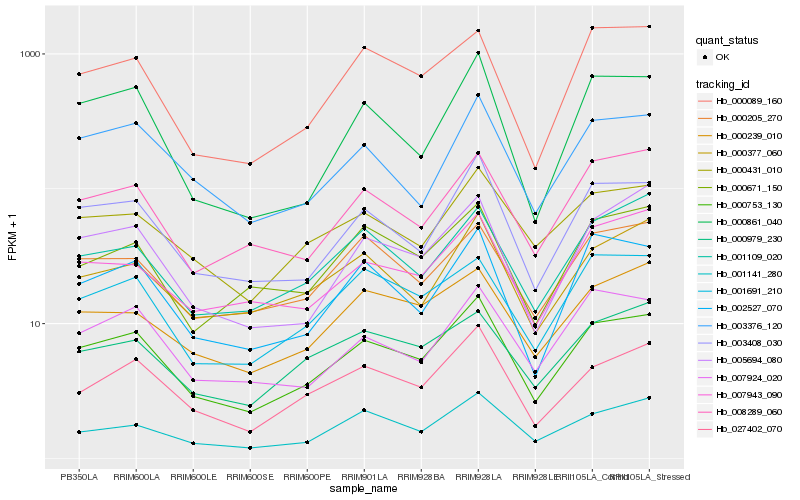
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000753_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640673 [Jatropha curcas] |
| 2 |
Hb_003408_030 |
0.0630084975 |
- |
- |
PREDICTED: protein YIF1B [Jatropha curcas] |
| 3 |
Hb_027402_070 |
0.0744066956 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 4 |
Hb_002527_070 |
0.0857275721 |
- |
- |
axonemal dynein light chain, putative [Ricinus communis] |
| 5 |
Hb_008289_060 |
0.0885196412 |
- |
- |
PREDICTED: general transcription factor IIE subunit 1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000431_010 |
0.0886236775 |
- |
- |
hypothetical protein POPTR_0016s05340g [Populus trichocarpa] |
| 7 |
Hb_001141_280 |
0.090490262 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g14820 [Vitis vinifera] |
| 8 |
Hb_001691_210 |
0.0912202625 |
transcription factor |
TF Family: C2C2-LSD |
hypothetical protein JCGZ_21436 [Jatropha curcas] |
| 9 |
Hb_000089_160 |
0.0915045616 |
- |
- |
60S ribosomal protein L5B [Hevea brasiliensis] |
| 10 |
Hb_003376_120 |
0.0924297527 |
- |
- |
PREDICTED: cell number regulator 8 [Jatropha curcas] |
| 11 |
Hb_000861_040 |
0.0938858785 |
- |
- |
PREDICTED: nascent polypeptide-associated complex subunit alpha-like protein 1 [Jatropha curcas] |
| 12 |
Hb_005694_080 |
0.0941678692 |
- |
- |
DNA-directed RNA polymerase III, putative [Ricinus communis] |
| 13 |
Hb_000377_060 |
0.0950548524 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 14 |
Hb_007924_020 |
0.0956900301 |
- |
- |
PREDICTED: chaperonin CPN60-like 2, mitochondrial isoform X2 [Jatropha curcas] |
| 15 |
Hb_000239_010 |
0.0959365968 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 16 |
Hb_000205_270 |
0.0960505515 |
- |
- |
PREDICTED: uncharacterized protein LOC105632722 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000671_150 |
0.097790975 |
- |
- |
PREDICTED: signal recognition particle 19 kDa protein-like [Jatropha curcas] |
| 18 |
Hb_001109_020 |
0.0986557178 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_007943_090 |
0.0986993908 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 20 |
Hb_000979_230 |
0.0994496191 |
- |
- |
PREDICTED: protein BCCIP homolog [Jatropha curcas] |