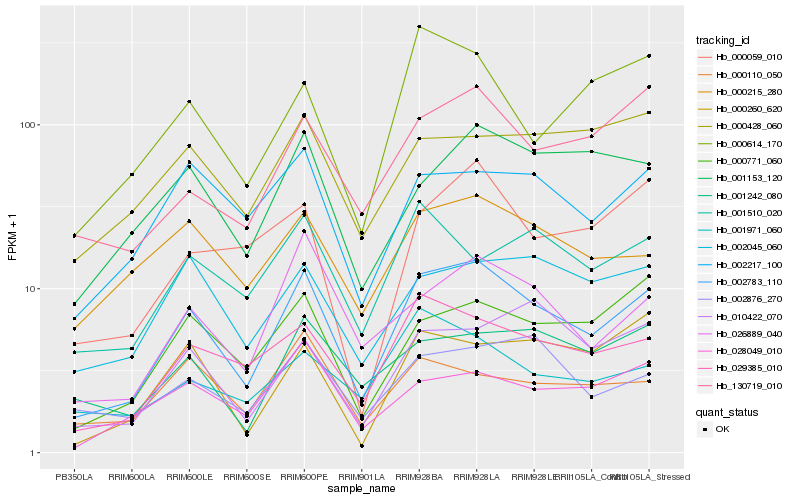
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002783_110 |
0.0 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 2 |
Hb_029385_010 |
0.1337924498 |
- |
- |
CTP synthase family protein [Populus trichocarpa] |
| 3 |
Hb_026889_040 |
0.1505967956 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 4 |
Hb_000771_060 |
0.1516906373 |
- |
- |
PREDICTED: spermine synthase [Jatropha curcas] |
| 5 |
Hb_002217_100 |
0.1551164992 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
dihydrolipoamide dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_130719_010 |
0.1585369833 |
- |
- |
V-type proton ATPase subunit E [Hevea brasiliensis] |
| 7 |
Hb_028049_010 |
0.1604055609 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000215_280 |
0.1616203777 |
- |
- |
Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor, putative [Ricinus communis] |
| 9 |
Hb_002045_060 |
0.16240718 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial [Jatropha curcas] |
| 10 |
Hb_002876_270 |
0.1631552332 |
- |
- |
PREDICTED: probable magnesium transporter NIPA4 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001153_120 |
0.1633862228 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 12 |
Hb_001242_080 |
0.1650907667 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Populus euphratica] |
| 13 |
Hb_000614_170 |
0.1703566863 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase-2C cytosolic [Gossypium arboreum] |
| 14 |
Hb_001510_020 |
0.1704250761 |
- |
- |
Diaminopimelate epimerase, putative [Ricinus communis] |
| 15 |
Hb_000059_010 |
0.1710778694 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_000260_620 |
0.1739524338 |
- |
- |
Thermosensitive gluconokinase, putative [Ricinus communis] |
| 17 |
Hb_000428_060 |
0.1741191233 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_000110_050 |
0.1743795311 |
- |
- |
- |
| 19 |
Hb_001971_060 |
0.1746896968 |
- |
- |
PREDICTED: 66 kDa stress protein-like [Populus euphratica] |
| 20 |
Hb_010422_070 |
0.1749146888 |
- |
- |
PREDICTED: UPF0483 protein CBG03338-like [Jatropha curcas] |