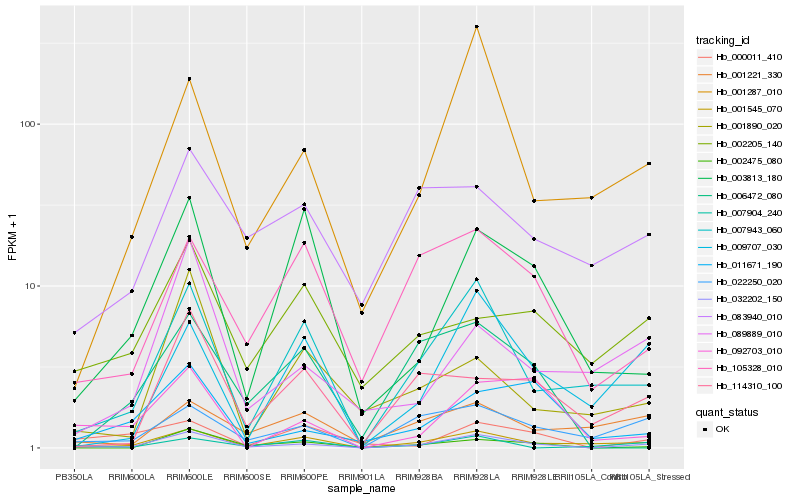
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032202_150 |
0.0 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR14 [Jatropha curcas] |
| 2 |
Hb_001545_070 |
0.2101347303 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_007943_060 |
0.2163596048 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 4 |
Hb_022250_020 |
0.2382289659 |
- |
- |
PREDICTED: uncharacterized protein LOC105648703 [Jatropha curcas] |
| 5 |
Hb_002475_080 |
0.2438774554 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein MALE MEIOCYTE DEATH 1 [Jatropha curcas] |
| 6 |
Hb_001287_010 |
0.254303294 |
- |
- |
PREDICTED: arginine decarboxylase [Jatropha curcas] |
| 7 |
Hb_009707_030 |
0.2562245532 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620 [Jatropha curcas] |
| 8 |
Hb_114310_100 |
0.2657745028 |
- |
- |
glucose-1-phosphate adenylyltransferase, putative [Ricinus communis] |
| 9 |
Hb_105328_010 |
0.2712251425 |
- |
- |
PREDICTED: low-temperature-induced cysteine proteinase-like [Jatropha curcas] |
| 10 |
Hb_011671_190 |
0.2757894875 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 11 |
Hb_092703_010 |
0.2782772979 |
- |
- |
PREDICTED: putative kinase-like protein TMKL1 [Jatropha curcas] |
| 12 |
Hb_089889_010 |
0.2824128695 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 [Jatropha curcas] |
| 13 |
Hb_003813_180 |
0.2899101524 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 14 |
Hb_083940_010 |
0.292524487 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 15 |
Hb_001221_330 |
0.2932619321 |
- |
- |
PREDICTED: uncharacterized protein LOC105648791 [Jatropha curcas] |
| 16 |
Hb_001890_020 |
0.2984296509 |
- |
- |
hypothetical protein CICLE_v10025198mg [Citrus clementina] |
| 17 |
Hb_002205_140 |
0.2987448406 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2b [Jatropha curcas] |
| 18 |
Hb_006472_080 |
0.2997538366 |
- |
- |
- |
| 19 |
Hb_000011_410 |
0.299889746 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007904_240 |
0.2999387613 |
- |
- |
PREDICTED: protein terminal ear1 [Jatropha curcas] |