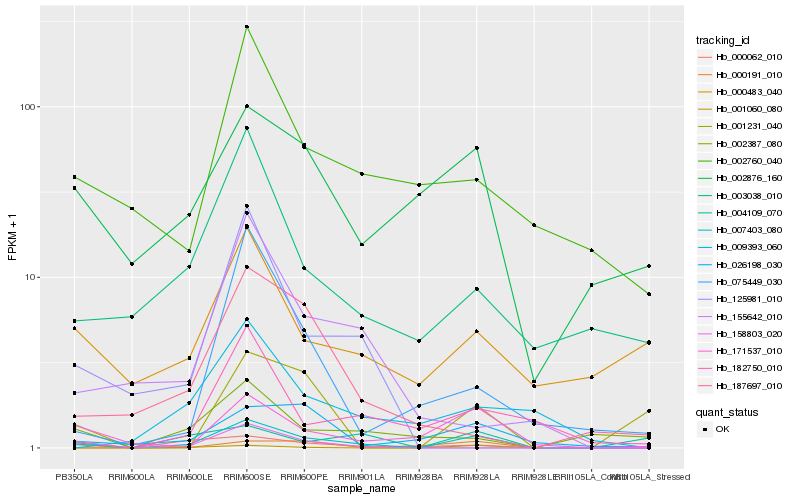
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007403_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000062_010 |
0.2929390123 |
- |
- |
Alpha-L-fucosidase 2 precursor, putative [Ricinus communis] |
| 3 |
Hb_171537_010 |
0.2988227356 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 4 |
Hb_001060_080 |
0.3107843555 |
- |
- |
PREDICTED: uncharacterized protein LOC104219776 [Nicotiana sylvestris] |
| 5 |
Hb_026198_030 |
0.3332375999 |
- |
- |
PREDICTED: ABC transporter G family member 25 [Jatropha curcas] |
| 6 |
Hb_182750_010 |
0.3336913506 |
- |
- |
hypothetical protein MIMGU_mgv1a0001602mg, partial [Erythranthe guttata] |
| 7 |
Hb_002876_160 |
0.3350135088 |
- |
- |
sucrose transporter 1 [Hevea brasiliensis] |
| 8 |
Hb_003038_010 |
0.3363948594 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 9 |
Hb_002760_040 |
0.3439434459 |
- |
- |
PREDICTED: uncharacterized protein LOC105641234 [Jatropha curcas] |
| 10 |
Hb_158803_020 |
0.3449151661 |
- |
- |
hypothetical protein JCGZ_12890 [Jatropha curcas] |
| 11 |
Hb_009393_060 |
0.3474551983 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 12 |
Hb_075449_030 |
0.3481163493 |
- |
- |
PREDICTED: two-pore potassium channel 1 [Jatropha curcas] |
| 13 |
Hb_155642_010 |
0.3495907197 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 14 |
Hb_001231_040 |
0.3549903599 |
- |
- |
hypothetical protein POPTR_0006s06470g [Populus trichocarpa] |
| 15 |
Hb_187697_010 |
0.3576783003 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 16 |
Hb_000191_010 |
0.3577859299 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 17 |
Hb_004109_070 |
0.3577978367 |
- |
- |
hypothetical protein PRUPE_ppa010934mg [Prunus persica] |
| 18 |
Hb_000483_040 |
0.3603213568 |
- |
- |
Cell division protein ftsH, putative [Ricinus communis] |
| 19 |
Hb_125981_010 |
0.3629483889 |
- |
- |
hypothetical protein POPTR_0005s042901g, partial [Populus trichocarpa] |
| 20 |
Hb_002387_080 |
0.3637373993 |
- |
- |
hypothetical protein JCGZ_25275 [Jatropha curcas] |