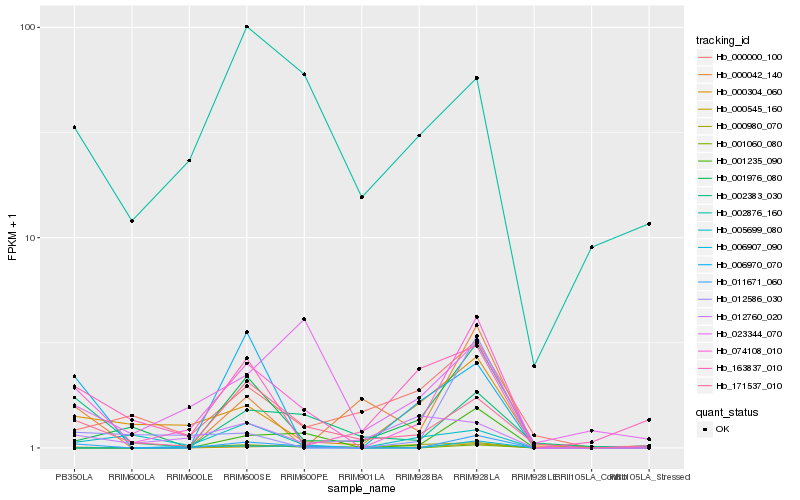
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_074108_010 |
0.0 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 2 |
Hb_011671_060 |
0.2128684692 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_26269 [Jatropha curcas] |
| 3 |
Hb_171537_010 |
0.2758260928 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 4 |
Hb_000304_060 |
0.288646596 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0010s15360g [Populus trichocarpa] |
| 5 |
Hb_002383_030 |
0.2971538151 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 6 |
Hb_000980_070 |
0.3250662455 |
- |
- |
PREDICTED: uncharacterized protein LOC103440883 [Malus domestica] |
| 7 |
Hb_001976_080 |
0.3280459816 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor AIL6 [Jatropha curcas] |
| 8 |
Hb_001235_090 |
0.3456698462 |
- |
- |
PREDICTED: uncharacterized protein LOC103449952 [Malus domestica] |
| 9 |
Hb_006970_070 |
0.3466901865 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001060_080 |
0.3585702413 |
- |
- |
PREDICTED: uncharacterized protein LOC104219776 [Nicotiana sylvestris] |
| 11 |
Hb_005699_080 |
0.3609138864 |
- |
- |
PREDICTED: ABC transporter G family member 10-like [Jatropha curcas] |
| 12 |
Hb_000000_100 |
0.3617235696 |
- |
- |
hypothetical protein POPTR_0002s13260g [Populus trichocarpa] |
| 13 |
Hb_012586_030 |
0.3628911086 |
- |
- |
hypothetical protein CICLE_v10014868mg [Citrus clementina] |
| 14 |
Hb_006907_090 |
0.3686136237 |
- |
- |
- |
| 15 |
Hb_163837_010 |
0.3719859719 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Eucalyptus grandis] |
| 16 |
Hb_000042_140 |
0.3720978433 |
- |
- |
hypothetical protein POPTR_0001s41640g, partial [Populus trichocarpa] |
| 17 |
Hb_002876_160 |
0.3738196918 |
- |
- |
sucrose transporter 1 [Hevea brasiliensis] |
| 18 |
Hb_000545_160 |
0.3743096711 |
- |
- |
PREDICTED: uncharacterized protein LOC105637052 [Jatropha curcas] |
| 19 |
Hb_023344_070 |
0.3863213621 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 20 |
Hb_012760_020 |
0.3928984191 |
- |
- |
PREDICTED: uncharacterized protein LOC105647080 [Jatropha curcas] |