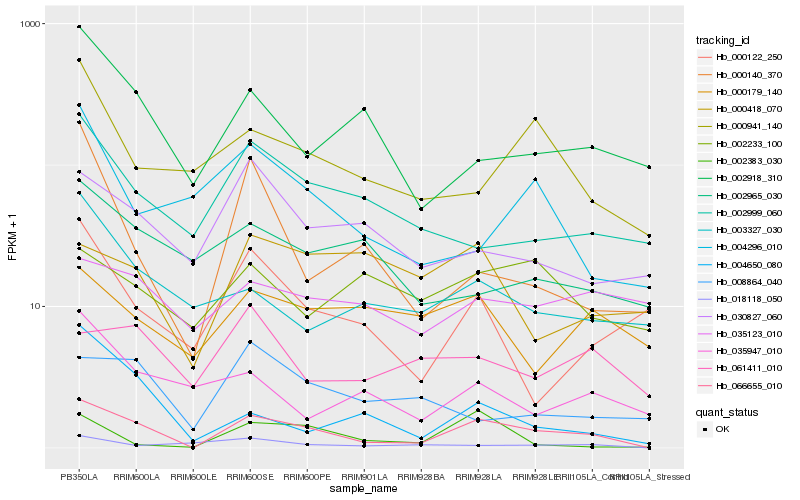
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_066655_010 |
0.0 |
- |
- |
PREDICTED: abnormal long morphology protein 1 isoform X3 [Jatropha curcas] |
| 2 |
Hb_002918_310 |
0.2953357135 |
- |
- |
PREDICTED: dnaJ protein homolog 1-like [Jatropha curcas] |
| 3 |
Hb_000941_140 |
0.2986737185 |
- |
- |
PREDICTED: activator of 90 kDa heat shock protein ATPase homolog 1 [Jatropha curcas] |
| 4 |
Hb_002999_060 |
0.2992119121 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 [Jatropha curcas] |
| 5 |
Hb_030827_060 |
0.3018165545 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor B-2b [Jatropha curcas] |
| 6 |
Hb_003327_030 |
0.3071787106 |
- |
- |
Hsc70-interacting protein, putative [Ricinus communis] |
| 7 |
Hb_000122_250 |
0.3077376071 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 2C, putative [Ricinus communis] |
| 8 |
Hb_004650_080 |
0.3082964144 |
- |
- |
- |
| 9 |
Hb_002383_030 |
0.3086645221 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 10 |
Hb_018118_050 |
0.3110147337 |
- |
- |
hypothetical protein POPTR_0006s07370g [Populus trichocarpa] |
| 11 |
Hb_008864_040 |
0.3113508851 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15980 [Jatropha curcas] |
| 12 |
Hb_004296_010 |
0.3115979216 |
- |
- |
PREDICTED: activator of 90 kDa heat shock protein ATPase homolog 1 [Jatropha curcas] |
| 13 |
Hb_061411_010 |
0.3116594576 |
- |
- |
PREDICTED: ATPase WRNIP1 [Jatropha curcas] |
| 14 |
Hb_000179_140 |
0.3137007071 |
- |
- |
phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 15 |
Hb_035947_010 |
0.314754005 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 16 |
Hb_002965_030 |
0.3161929858 |
- |
- |
PREDICTED: uncharacterized protein LOC105643493 [Jatropha curcas] |
| 17 |
Hb_035123_010 |
0.3166164058 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 18 |
Hb_002233_100 |
0.3186997083 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 6-like [Malus domestica] |
| 19 |
Hb_000140_370 |
0.3190711613 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 6 [Jatropha curcas] |
| 20 |
Hb_000418_070 |
0.3201130429 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |