| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
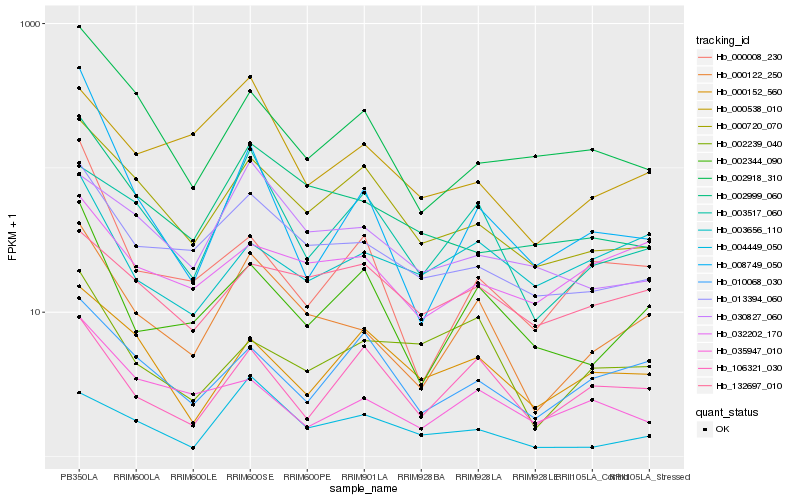
Hb_000122_250 |
0.0 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 2C, putative [Ricinus communis] |
| 2 |
Hb_013394_060 |
0.159674456 |
- |
- |
PREDICTED: IST1-like protein isoform X1 [Jatropha curcas] |
| 3 |
Hb_002344_090 |
0.1711581205 |
- |
- |
PREDICTED: uncharacterized protein LOC105637976 [Jatropha curcas] |
| 4 |
Hb_002999_060 |
0.179556936 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 [Jatropha curcas] |
| 5 |
Hb_000720_070 |
0.1882454296 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_20639 [Jatropha curcas] |
| 6 |
Hb_003517_060 |
0.1894491412 |
- |
- |
hypothetical protein CISIN_1g044999mg, partial [Citrus sinensis] |
| 7 |
Hb_000538_010 |
0.2033717334 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_030827_060 |
0.2043993706 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor B-2b [Jatropha curcas] |
| 9 |
Hb_002918_310 |
0.2046636071 |
- |
- |
PREDICTED: dnaJ protein homolog 1-like [Jatropha curcas] |
| 10 |
Hb_004449_050 |
0.2051099009 |
- |
- |
PREDICTED: tyrosine-sulfated glycopeptide receptor 1-like [Jatropha curcas] |
| 11 |
Hb_035947_010 |
0.2085605242 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 12 |
Hb_106321_030 |
0.2092428308 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 13 |
Hb_032202_170 |
0.2092774932 |
- |
- |
PREDICTED: uncharacterized protein LOC105643056 [Jatropha curcas] |
| 14 |
Hb_010068_030 |
0.209538782 |
- |
- |
aconitase, putative [Ricinus communis] |
| 15 |
Hb_000008_230 |
0.2161878958 |
- |
- |
hypothetical protein POPTR_0004s22120g [Populus trichocarpa] |
| 16 |
Hb_008749_050 |
0.2163381063 |
- |
- |
PREDICTED: uncharacterized J domain-containing protein C4H3.01 [Jatropha curcas] |
| 17 |
Hb_002239_040 |
0.216809244 |
- |
- |
Cellular apoptosis susceptibility protein / importin-alpha re-exporter, putative isoform 1 [Theobroma cacao] |
| 18 |
Hb_003656_110 |
0.217385584 |
- |
- |
phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 19 |
Hb_132697_010 |
0.219132788 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000152_560 |
0.2219049948 |
- |
- |
hypothetical protein JCGZ_26236 [Jatropha curcas] |