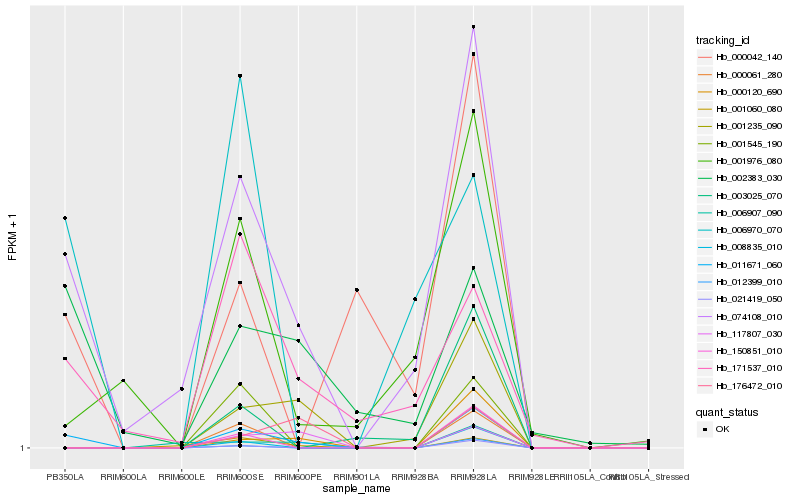
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011671_060 |
0.0 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_26269 [Jatropha curcas] |
| 2 |
Hb_074108_010 |
0.2128684692 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 3 |
Hb_002383_030 |
0.3325090622 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 4 |
Hb_171537_010 |
0.3438717965 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 5 |
Hb_000120_690 |
0.3512489442 |
- |
- |
PREDICTED: uncharacterized protein LOC105641027 [Jatropha curcas] |
| 6 |
Hb_001235_090 |
0.3582336081 |
- |
- |
PREDICTED: uncharacterized protein LOC103449952 [Malus domestica] |
| 7 |
Hb_150851_010 |
0.3591519653 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 8 |
Hb_000061_280 |
0.3600962452 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g44230 [Jatropha curcas] |
| 9 |
Hb_008835_010 |
0.3613539451 |
- |
- |
PREDICTED: uncharacterized protein LOC105851721 [Cicer arietinum] |
| 10 |
Hb_000042_140 |
0.3677561848 |
- |
- |
hypothetical protein POPTR_0001s41640g, partial [Populus trichocarpa] |
| 11 |
Hb_012399_010 |
0.3700583441 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_001545_190 |
0.377823483 |
- |
- |
hypothetical protein VITISV_012031 [Vitis vinifera] |
| 13 |
Hb_006970_070 |
0.3817357577 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_117807_030 |
0.3852419663 |
- |
- |
Protein MCM10 [Gossypium arboreum] |
| 15 |
Hb_001060_080 |
0.3855330223 |
- |
- |
PREDICTED: uncharacterized protein LOC104219776 [Nicotiana sylvestris] |
| 16 |
Hb_001976_080 |
0.3858722153 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor AIL6 [Jatropha curcas] |
| 17 |
Hb_006907_090 |
0.4054655213 |
- |
- |
- |
| 18 |
Hb_003025_070 |
0.408668347 |
- |
- |
PREDICTED: uncharacterized protein LOC103419743 [Malus domestica] |
| 19 |
Hb_176472_010 |
0.4127605477 |
- |
- |
BnaC01g33060D [Brassica napus] |
| 20 |
Hb_021419_050 |
0.4138859969 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Malus domestica] |