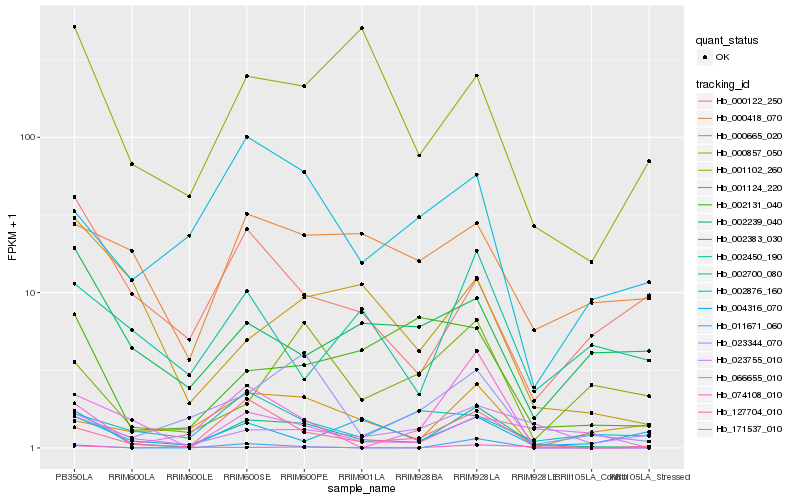
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002383_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 2 |
Hb_171537_010 |
0.2355770518 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 3 |
Hb_074108_010 |
0.2971538151 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 4 |
Hb_023755_010 |
0.3073397218 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 5 |
Hb_066655_010 |
0.3086645221 |
- |
- |
PREDICTED: abnormal long morphology protein 1 isoform X3 [Jatropha curcas] |
| 6 |
Hb_000418_070 |
0.3173607995 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 7 |
Hb_002876_160 |
0.3195493318 |
- |
- |
sucrose transporter 1 [Hevea brasiliensis] |
| 8 |
Hb_001102_260 |
0.3239136722 |
- |
- |
hypothetical protein L484_019841 [Morus notabilis] |
| 9 |
Hb_002450_190 |
0.3258717839 |
- |
- |
Nonspecific lipid-transfer protein precursor, putative [Ricinus communis] |
| 10 |
Hb_000665_020 |
0.3262109814 |
- |
- |
- |
| 11 |
Hb_002131_040 |
0.3299147935 |
- |
- |
proton-dependent oligopeptide transport family protein [Populus trichocarpa] |
| 12 |
Hb_011671_060 |
0.3325090622 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_26269 [Jatropha curcas] |
| 13 |
Hb_000857_050 |
0.3345229532 |
- |
- |
PREDICTED: protein YLS9 [Jatropha curcas] |
| 14 |
Hb_000122_250 |
0.3374726158 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 2C, putative [Ricinus communis] |
| 15 |
Hb_023344_070 |
0.3395487532 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 16 |
Hb_004316_070 |
0.3406823444 |
- |
- |
E3 ubiquitin-protein ligase UPL5 -like protein [Gossypium arboreum] |
| 17 |
Hb_127704_010 |
0.3416245489 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_002239_040 |
0.3436010814 |
- |
- |
Cellular apoptosis susceptibility protein / importin-alpha re-exporter, putative isoform 1 [Theobroma cacao] |
| 19 |
Hb_001124_220 |
0.3453403149 |
- |
- |
DsRNA-binding protein 5, putative isoform 1 [Theobroma cacao] |
| 20 |
Hb_002700_080 |
0.3495078705 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_24521 [Jatropha curcas] |