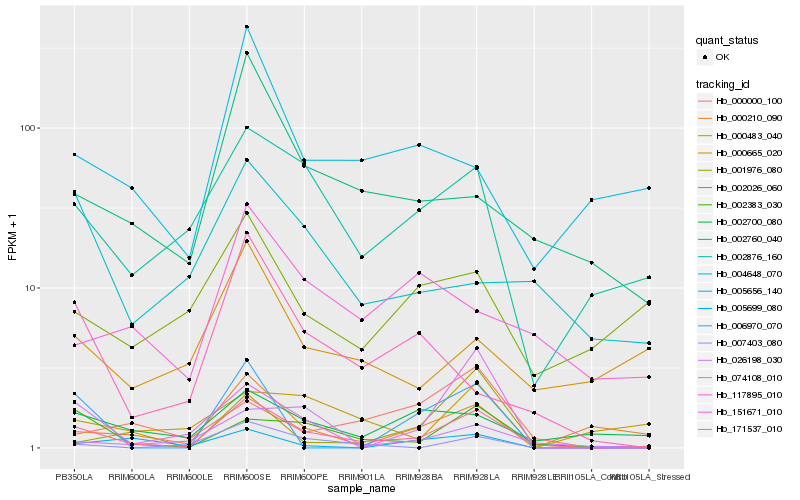
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_171537_010 |
0.0 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 2 |
Hb_002383_030 |
0.2355770518 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 3 |
Hb_002876_160 |
0.2551726071 |
- |
- |
sucrose transporter 1 [Hevea brasiliensis] |
| 4 |
Hb_074108_010 |
0.2758260928 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 5 |
Hb_000210_090 |
0.2909929655 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG7 [Jatropha curcas] |
| 6 |
Hb_002760_040 |
0.2917305114 |
- |
- |
PREDICTED: uncharacterized protein LOC105641234 [Jatropha curcas] |
| 7 |
Hb_002700_080 |
0.2919533306 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_24521 [Jatropha curcas] |
| 8 |
Hb_007403_080 |
0.2988227356 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000483_040 |
0.3145173001 |
- |
- |
Cell division protein ftsH, putative [Ricinus communis] |
| 10 |
Hb_002026_060 |
0.3148918393 |
- |
- |
PREDICTED: uncharacterized protein LOC105645786 [Jatropha curcas] |
| 11 |
Hb_026198_030 |
0.3158292501 |
- |
- |
PREDICTED: ABC transporter G family member 25 [Jatropha curcas] |
| 12 |
Hb_005656_140 |
0.3159266447 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 9 [Jatropha curcas] |
| 13 |
Hb_151671_010 |
0.3176080221 |
- |
- |
hypothetical protein POPTR_0005s04441g [Populus trichocarpa] |
| 14 |
Hb_001976_080 |
0.3182725464 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor AIL6 [Jatropha curcas] |
| 15 |
Hb_117895_010 |
0.3194164928 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |
| 16 |
Hb_004648_070 |
0.3201595994 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 17 |
Hb_006970_070 |
0.3225622383 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005699_080 |
0.3302871174 |
- |
- |
PREDICTED: ABC transporter G family member 10-like [Jatropha curcas] |
| 19 |
Hb_000000_100 |
0.3321734985 |
- |
- |
hypothetical protein POPTR_0002s13260g [Populus trichocarpa] |
| 20 |
Hb_000665_020 |
0.3339545689 |
- |
- |
- |