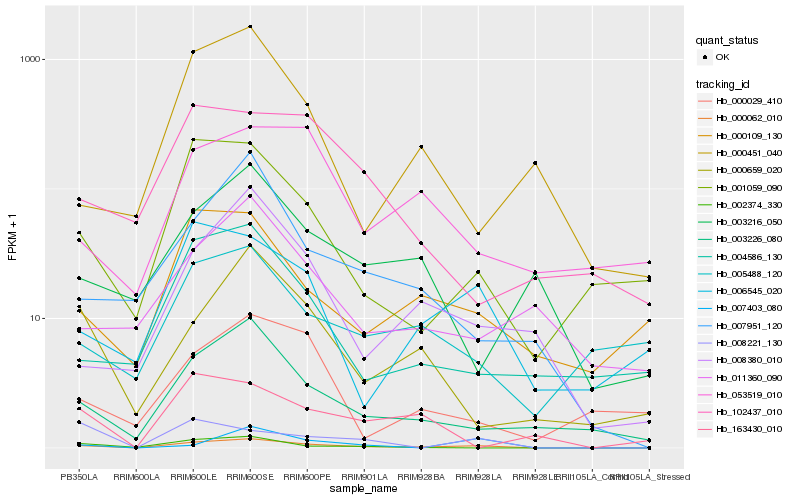
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000062_010 |
0.0 |
- |
- |
Alpha-L-fucosidase 2 precursor, putative [Ricinus communis] |
| 2 |
Hb_003226_080 |
0.2297028928 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5-like [Jatropha curcas] |
| 3 |
Hb_001059_090 |
0.2594259102 |
- |
- |
PREDICTED: F-box protein At1g61340 [Jatropha curcas] |
| 4 |
Hb_006545_020 |
0.268895 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 5 |
Hb_008380_010 |
0.2693991879 |
- |
- |
PREDICTED: PRA1 family protein B4-like [Camelina sativa] |
| 6 |
Hb_007951_120 |
0.2734443983 |
- |
- |
PREDICTED: uncharacterized protein At1g15400 [Populus euphratica] |
| 7 |
Hb_000109_130 |
0.2742515621 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 9 [Jatropha curcas] |
| 8 |
Hb_000659_020 |
0.2750189203 |
- |
- |
PREDICTED: putative L-type lectin-domain containing receptor kinase II.2 [Jatropha curcas] |
| 9 |
Hb_002374_330 |
0.277224475 |
- |
- |
- |
| 10 |
Hb_004586_130 |
0.286098274 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005488_120 |
0.2927843107 |
- |
- |
PREDICTED: DNA polymerase delta subunit 4 [Jatropha curcas] |
| 12 |
Hb_007403_080 |
0.2929390123 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_008221_130 |
0.2946426826 |
- |
- |
hypothetical protein VITISV_016971 [Vitis vinifera] |
| 14 |
Hb_053519_010 |
0.2961187653 |
- |
- |
PREDICTED: probable zinc transporter protein DDB_G0282067 [Jatropha curcas] |
| 15 |
Hb_102437_010 |
0.2982422459 |
- |
- |
PREDICTED: B-cell receptor-associated protein 31 [Beta vulgaris subsp. vulgaris] |
| 16 |
Hb_011360_090 |
0.2983176856 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 17 |
Hb_000029_410 |
0.2985163994 |
- |
- |
AT14A, putative [Ricinus communis] |
| 18 |
Hb_000451_040 |
0.3009818174 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_163430_010 |
0.3040704177 |
- |
- |
Polyneuridine-aldehyde esterase precursor, putative [Ricinus communis] |
| 20 |
Hb_003216_050 |
0.3048579864 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |