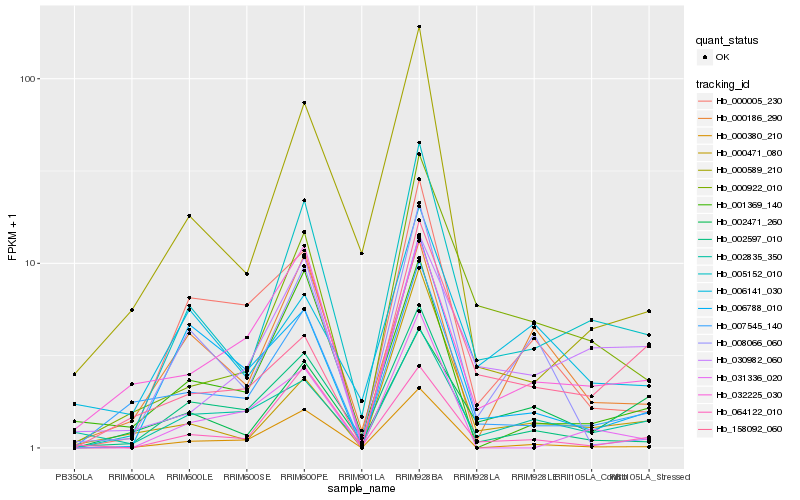
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005152_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_22048 [Jatropha curcas] |
| 2 |
Hb_007545_140 |
0.1692881747 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105141503 [Populus euphratica] |
| 3 |
Hb_000922_010 |
0.1711158904 |
- |
- |
hypothetical protein JCGZ_11488 [Jatropha curcas] |
| 4 |
Hb_002597_010 |
0.1847761489 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Malus domestica] |
| 5 |
Hb_064122_010 |
0.1999418802 |
- |
- |
PREDICTED: uncharacterized protein LOC105631858 isoform X2 [Jatropha curcas] |
| 6 |
Hb_030982_060 |
0.2020867916 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002471_260 |
0.2029854754 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105634154 [Jatropha curcas] |
| 8 |
Hb_000589_210 |
0.2030406949 |
- |
- |
PREDICTED: uncharacterized protein LOC105647439 [Jatropha curcas] |
| 9 |
Hb_002835_350 |
0.2063585084 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 10 |
Hb_006788_010 |
0.2067110342 |
- |
- |
PREDICTED: calcium-transporting ATPase 12, plasma membrane-type [Jatropha curcas] |
| 11 |
Hb_000005_230 |
0.2105999114 |
- |
- |
hypothetical protein POPTR_0009s01160g [Populus trichocarpa] |
| 12 |
Hb_000471_080 |
0.2107002685 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 13 |
Hb_000380_210 |
0.2107642498 |
- |
- |
Cyclic nucleotide-gated channel 15 [Theobroma cacao] |
| 14 |
Hb_001369_140 |
0.2167063829 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_158092_060 |
0.2197553962 |
- |
- |
aspartate semialdehyde dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_008066_060 |
0.2208020028 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 17 |
Hb_000186_290 |
0.2231824175 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 8 isoform X1 [Jatropha curcas] |
| 18 |
Hb_006141_030 |
0.2253506546 |
- |
- |
- |
| 19 |
Hb_032225_030 |
0.2275366244 |
- |
- |
PREDICTED: leucine-rich repeat extensin-like protein 2 [Jatropha curcas] |
| 20 |
Hb_031336_020 |
0.2278593082 |
- |
- |
conserved hypothetical protein [Ricinus communis] |