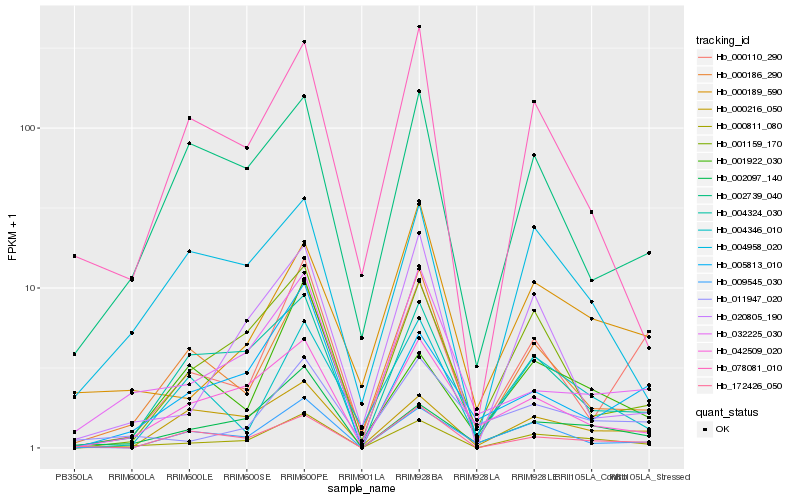
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000811_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002097_140 |
0.1706437557 |
- |
- |
CHY zinc finger family protein, expressed [Oryza sativa Japonica Group] |
| 3 |
Hb_001159_170 |
0.1897990787 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 4 |
Hb_004324_030 |
0.1897991611 |
- |
- |
PREDICTED: probable protein phosphatase 2C 8 [Jatropha curcas] |
| 5 |
Hb_000186_290 |
0.1945657843 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 8 isoform X1 [Jatropha curcas] |
| 6 |
Hb_042509_020 |
0.1979760375 |
- |
- |
hypothetical protein JCGZ_19555 [Jatropha curcas] |
| 7 |
Hb_009545_030 |
0.2044905935 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_004958_020 |
0.2086515261 |
- |
- |
PREDICTED: uncharacterized protein LOC104609039 [Nelumbo nucifera] |
| 9 |
Hb_000216_050 |
0.2133657345 |
- |
- |
PREDICTED: indole-3-acetaldehyde oxidase-like [Jatropha curcas] |
| 10 |
Hb_172426_050 |
0.2161622503 |
- |
- |
PREDICTED: uncharacterized protein LOC105634266 [Jatropha curcas] |
| 11 |
Hb_032225_030 |
0.2178489844 |
- |
- |
PREDICTED: leucine-rich repeat extensin-like protein 2 [Jatropha curcas] |
| 12 |
Hb_020805_190 |
0.2185911664 |
transcription factor |
TF Family: HB |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_001922_030 |
0.2192148824 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 5 [Jatropha curcas] |
| 14 |
Hb_004346_010 |
0.2196660674 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005813_010 |
0.2197591385 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_078081_010 |
0.2217548623 |
- |
- |
hypothetical protein PRUPE_ppa015726mg [Prunus persica] |
| 17 |
Hb_011947_020 |
0.225575476 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 18 |
Hb_000110_290 |
0.2258321857 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002739_040 |
0.2261473598 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 20 |
Hb_000189_590 |
0.2279579344 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |