| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
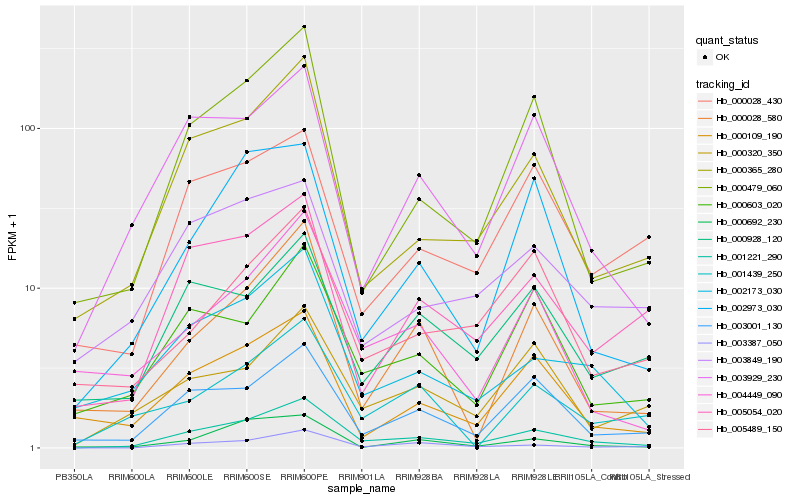
Hb_001221_290 |
0.0 |
- |
- |
hypothetical protein RCOM_1506010 [Ricinus communis] |
| 2 |
Hb_000365_280 |
0.1392228186 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |
| 3 |
Hb_003001_130 |
0.1508431363 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002173_030 |
0.1517539546 |
- |
- |
hypothetical protein JCGZ_12466 [Jatropha curcas] |
| 5 |
Hb_000479_060 |
0.1535007771 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1 [Vitis vinifera] |
| 6 |
Hb_003387_050 |
0.1543722635 |
- |
- |
PREDICTED: uncharacterized protein LOC105639324 [Jatropha curcas] |
| 7 |
Hb_003929_230 |
0.164181951 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 5 [Nelumbo nucifera] |
| 8 |
Hb_005054_020 |
0.1664205577 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 9 |
Hb_000603_020 |
0.1721602479 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000320_350 |
0.1736261999 |
- |
- |
hypothetical protein RCOM_0904330 [Ricinus communis] |
| 11 |
Hb_000109_190 |
0.1768458131 |
- |
- |
PREDICTED: RING-H2 finger protein ATL47 [Jatropha curcas] |
| 12 |
Hb_004449_090 |
0.1771665022 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 13 |
Hb_002973_030 |
0.1796224952 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 14 |
Hb_000028_580 |
0.1801872969 |
- |
- |
PREDICTED: glutamate decarboxylase 5-like [Jatropha curcas] |
| 15 |
Hb_000928_120 |
0.1803556522 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 16 |
Hb_000028_430 |
0.1830178035 |
- |
- |
PREDICTED: uncharacterized protein LOC105641369 isoform X1 [Jatropha curcas] |
| 17 |
Hb_005489_150 |
0.1857458608 |
- |
- |
hypothetical protein JCGZ_20759 [Jatropha curcas] |
| 18 |
Hb_000692_230 |
0.1871746422 |
- |
- |
phospholipase d delta, putative [Ricinus communis] |
| 19 |
Hb_003849_190 |
0.1879287223 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 20 |
Hb_001439_250 |
0.188618634 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100 [Jatropha curcas] |