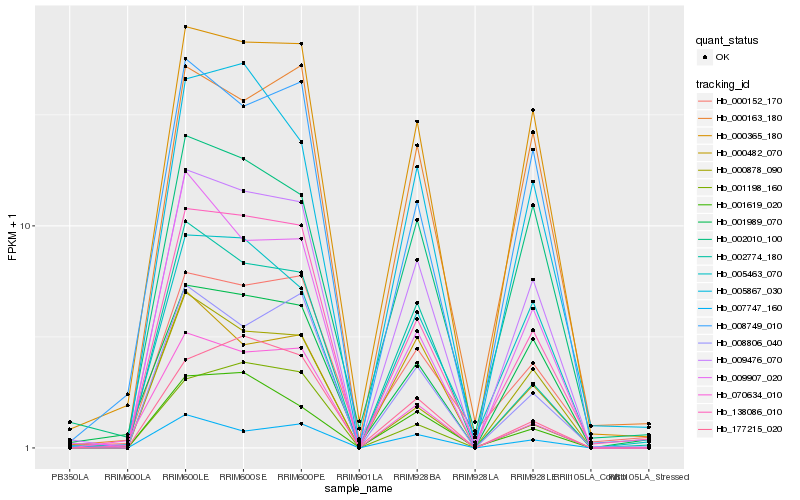
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009476_070 |
0.0 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 2 |
Hb_000365_180 |
0.0691036828 |
- |
- |
PREDICTED: protein LURP-one-related 8-like [Pyrus x bretschneideri] |
| 3 |
Hb_138086_010 |
0.0944448382 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 4 |
Hb_008806_040 |
0.1099262695 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 5 |
Hb_008749_010 |
0.110939485 |
- |
- |
PREDICTED: auxin efflux carrier component 3-like [Jatropha curcas] |
| 6 |
Hb_002774_180 |
0.1121116574 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 7 |
Hb_005463_070 |
0.1144158173 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 8 |
Hb_000163_180 |
0.1151084981 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |
| 9 |
Hb_005867_030 |
0.1160500281 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 10 |
Hb_002010_100 |
0.1162604582 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 11 |
Hb_001989_070 |
0.1194778672 |
- |
- |
PREDICTED: uncharacterized calcium-binding protein At1g02270 [Jatropha curcas] |
| 12 |
Hb_000878_090 |
0.1196370691 |
- |
- |
sucrose-phosphatase family protein [Populus trichocarpa] |
| 13 |
Hb_001619_020 |
0.1210160881 |
- |
- |
LRR receptor-like kinase family protein [Medicago truncatula] |
| 14 |
Hb_007747_160 |
0.1221622181 |
- |
- |
lysosomal pro-X carboxypeptidase, putative [Ricinus communis] |
| 15 |
Hb_009907_020 |
0.1231121783 |
- |
- |
hypothetical protein CICLE_v10009323mg [Citrus clementina] |
| 16 |
Hb_000482_070 |
0.1236354986 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 17 |
Hb_070634_010 |
0.1282888578 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 18 |
Hb_177215_020 |
0.1294991897 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 19 |
Hb_000152_170 |
0.1319073355 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |
| 20 |
Hb_001198_160 |
0.1326596054 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |