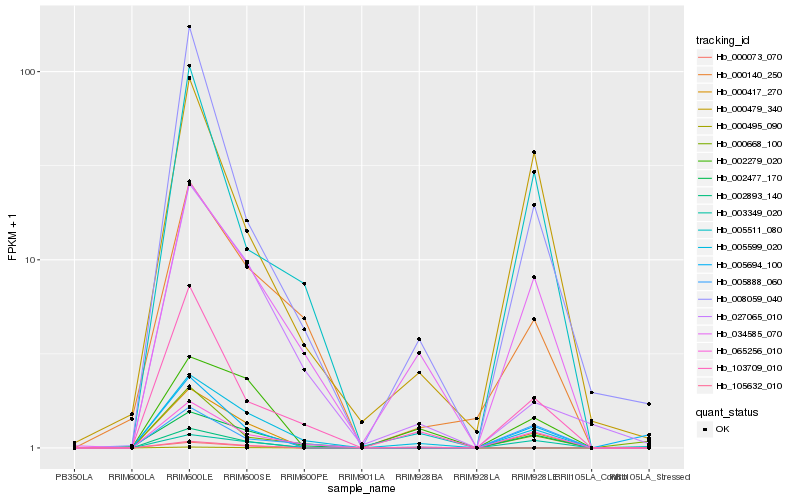
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000417_270 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002477_170 |
0.1634461675 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_065256_010 |
0.1771955258 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 4 |
Hb_005599_020 |
0.1803149494 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 5 |
Hb_003349_020 |
0.2165255586 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Beta vulgaris subsp. vulgaris] |
| 6 |
Hb_002279_020 |
0.2203443413 |
- |
- |
hypothetical protein POPTR_0009s03100g [Populus trichocarpa] |
| 7 |
Hb_005694_100 |
0.2245642205 |
- |
- |
PREDICTED: calcium-binding protein KIC-like [Populus euphratica] |
| 8 |
Hb_008059_040 |
0.2318278945 |
- |
- |
hypothetical protein POPTR_0019s12360g [Populus trichocarpa] |
| 9 |
Hb_000479_340 |
0.2330536815 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 10 |
Hb_103709_010 |
0.2338460496 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 11 |
Hb_034585_070 |
0.2342628613 |
- |
- |
Polyneuridine-aldehyde esterase precursor, putative [Ricinus communis] |
| 12 |
Hb_005511_080 |
0.2374001535 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 13 |
Hb_027065_010 |
0.2506087154 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 14 |
Hb_000140_250 |
0.2512377962 |
- |
- |
Late embryogenesis abundant protein Lea14-A, putative [Ricinus communis] |
| 15 |
Hb_005888_060 |
0.2515668853 |
- |
- |
hypothetical protein PRUPE_ppa002432m2g, partial [Prunus persica] |
| 16 |
Hb_000668_100 |
0.2535395773 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 17 |
Hb_000073_070 |
0.2546867055 |
- |
- |
- |
| 18 |
Hb_000495_090 |
0.2547353217 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 19 |
Hb_002893_140 |
0.2547913129 |
- |
- |
Glycine-rich cell wall structural protein precursor, putative [Ricinus communis] |
| 20 |
Hb_105632_010 |
0.2550860861 |
- |
- |
- |