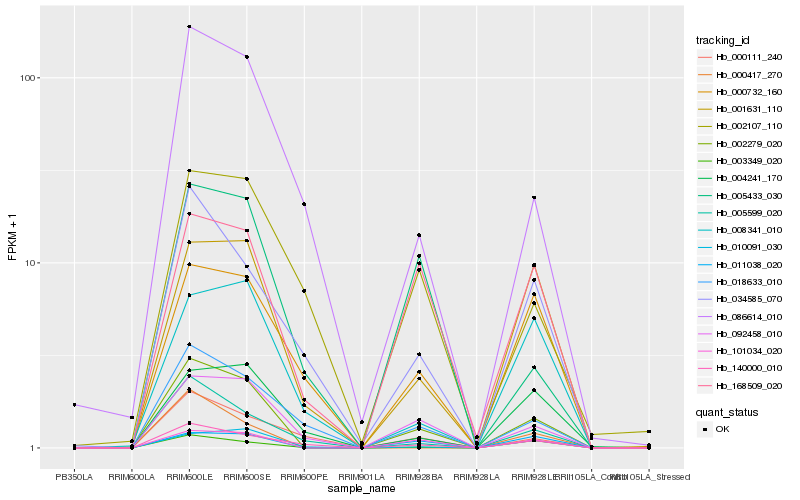
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002279_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0009s03100g [Populus trichocarpa] |
| 2 |
Hb_101034_020 |
0.1606601329 |
- |
- |
hypothetical protein JCGZ_13470 [Jatropha curcas] |
| 3 |
Hb_092458_010 |
0.1716670179 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 4 |
Hb_018633_010 |
0.1895717612 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 5 |
Hb_140000_010 |
0.1920812931 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 6 |
Hb_005599_020 |
0.1924490992 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 7 |
Hb_086614_010 |
0.1949537686 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein HD1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_034585_070 |
0.2073247734 |
- |
- |
Polyneuridine-aldehyde esterase precursor, putative [Ricinus communis] |
| 9 |
Hb_011038_020 |
0.207551011 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001631_110 |
0.2118867451 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: gamma-glutamyltranspeptidase 1-like [Populus euphratica] |
| 11 |
Hb_010091_030 |
0.2125376367 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 12 |
Hb_003349_020 |
0.2138391076 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Beta vulgaris subsp. vulgaris] |
| 13 |
Hb_168509_020 |
0.2147928732 |
- |
- |
macrophage migration inhibitory factor family protein [Populus trichocarpa] |
| 14 |
Hb_000111_240 |
0.2202296703 |
- |
- |
PREDICTED: glutamate receptor 2.8-like [Elaeis guineensis] |
| 15 |
Hb_000417_270 |
0.2203443413 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005433_030 |
0.222895876 |
transcription factor |
TF Family: MYB |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_000732_160 |
0.2323723227 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 18 |
Hb_002107_110 |
0.2361160537 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 6 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004241_170 |
0.237924442 |
- |
- |
PREDICTED: uncharacterized protein LOC105633220 [Jatropha curcas] |
| 20 |
Hb_008341_010 |
0.2405040069 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein HD1-like isoform X2 [Jatropha curcas] |