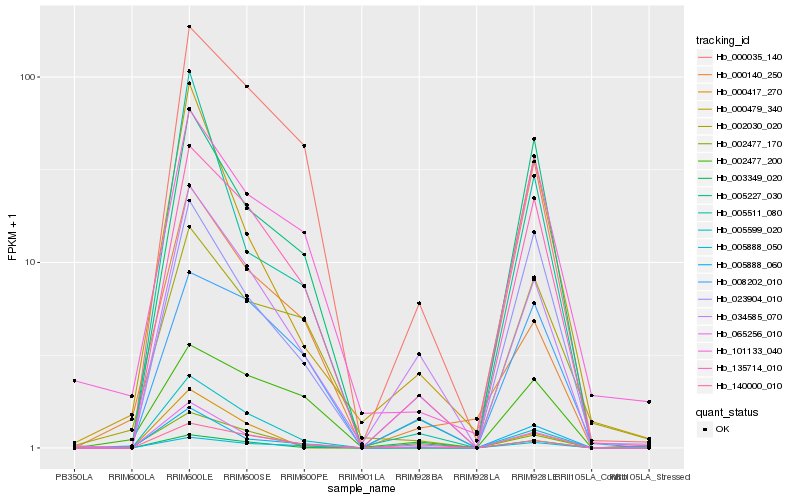
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002477_170 |
0.0 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_065256_010 |
0.148158032 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 3 |
Hb_000417_270 |
0.1634461675 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005599_020 |
0.1649379295 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 5 |
Hb_135714_010 |
0.1690928987 |
- |
- |
hypothetical protein POPTR_0017s09310g, partial [Populus trichocarpa] |
| 6 |
Hb_003349_020 |
0.1736690039 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Beta vulgaris subsp. vulgaris] |
| 7 |
Hb_000140_250 |
0.1868905006 |
- |
- |
Late embryogenesis abundant protein Lea14-A, putative [Ricinus communis] |
| 8 |
Hb_034585_070 |
0.1940528126 |
- |
- |
Polyneuridine-aldehyde esterase precursor, putative [Ricinus communis] |
| 9 |
Hb_005888_060 |
0.1973106177 |
- |
- |
hypothetical protein PRUPE_ppa002432m2g, partial [Prunus persica] |
| 10 |
Hb_005888_050 |
0.2032832382 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_000479_340 |
0.207229095 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 12 |
Hb_140000_010 |
0.207448743 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 13 |
Hb_101133_040 |
0.2152985272 |
- |
- |
PREDICTED: nudix hydrolase 18, mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_000035_140 |
0.2170083116 |
transcription factor |
TF Family: WRKY |
putative WRKY transcription factor family protein [Populus trichocarpa] |
| 15 |
Hb_005227_030 |
0.2170908416 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP28, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002477_200 |
0.2223837652 |
- |
- |
PREDICTED: uncharacterized protein LOC105631678 [Jatropha curcas] |
| 17 |
Hb_002030_020 |
0.2230016007 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2-like [Jatropha curcas] |
| 18 |
Hb_023904_010 |
0.2231226936 |
- |
- |
- |
| 19 |
Hb_005511_080 |
0.2268857511 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 20 |
Hb_008202_010 |
0.2284920516 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |