| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
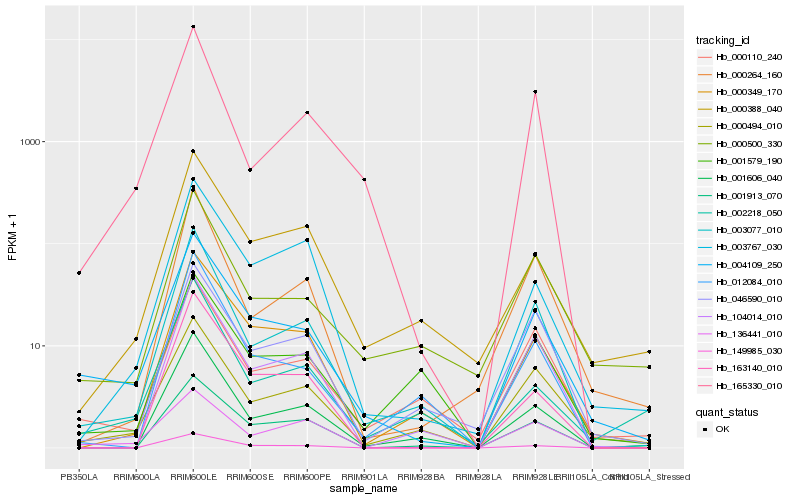
Hb_004109_250 |
0.0 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 2 |
Hb_003077_010 |
0.1478694998 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit O, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_000494_010 |
0.1518466781 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0014s04460g [Populus trichocarpa] |
| 4 |
Hb_000349_170 |
0.1536063206 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 5 |
Hb_000110_240 |
0.1652890703 |
- |
- |
PREDICTED: uncharacterized protein LOC105642006 isoform X3 [Jatropha curcas] |
| 6 |
Hb_000500_330 |
0.1662924084 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET2a-like [Jatropha curcas] |
| 7 |
Hb_000388_040 |
0.1693684104 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 8 |
Hb_003767_030 |
0.1785302556 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 9 |
Hb_165330_010 |
0.1795935406 |
- |
- |
BnaUnng00840D [Brassica napus] |
| 10 |
Hb_104014_010 |
0.1829877687 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39020 isoform X2 [Jatropha curcas] |
| 11 |
Hb_136441_010 |
0.184108142 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 12 |
Hb_163140_010 |
0.1841505439 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 13 |
Hb_149985_030 |
0.1842116743 |
- |
- |
PREDICTED: zeatin O-glucosyltransferase-like [Jatropha curcas] |
| 14 |
Hb_000264_160 |
0.185656183 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX5-like [Jatropha curcas] |
| 15 |
Hb_012084_010 |
0.1870015266 |
- |
- |
cold regulated protein [Manihot esculenta] |
| 16 |
Hb_046590_010 |
0.1872023334 |
- |
- |
Cycloeucalenol cycloisomerase, putative [Ricinus communis] |
| 17 |
Hb_001606_040 |
0.1880795465 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 18 |
Hb_001579_190 |
0.1881478723 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002218_050 |
0.188313174 |
- |
- |
SSXT family protein isoform 2, partial [Theobroma cacao] |
| 20 |
Hb_001913_070 |
0.1884408649 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |