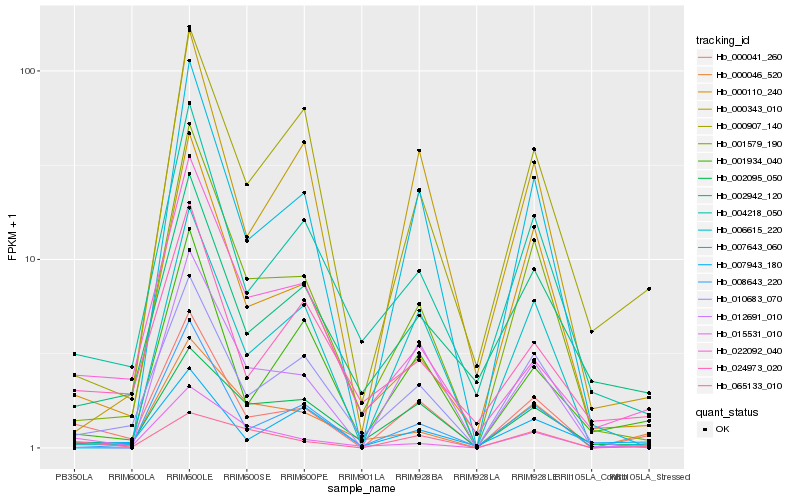
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000041_260 |
0.0 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 2 |
Hb_001579_190 |
0.1876658188 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004218_050 |
0.1880529557 |
- |
- |
annexin [Manihot esculenta] |
| 4 |
Hb_001934_040 |
0.1895849031 |
- |
- |
PREDICTED: vacuolar iron transporter 1-like [Jatropha curcas] |
| 5 |
Hb_010683_070 |
0.1920232033 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase CMT2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_006615_220 |
0.1997816097 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BIM1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000343_010 |
0.2000144948 |
- |
- |
F3F9.11 [Arabidopsis thaliana] |
| 8 |
Hb_024973_020 |
0.2000765548 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 9 |
Hb_015531_010 |
0.2012753187 |
- |
- |
PREDICTED: uncharacterized protein LOC105636764 [Jatropha curcas] |
| 10 |
Hb_008643_220 |
0.2017505975 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 [Jatropha curcas] |
| 11 |
Hb_022092_040 |
0.20529797 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 [Jatropha curcas] |
| 12 |
Hb_000046_520 |
0.2072218383 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000110_240 |
0.2076039302 |
- |
- |
PREDICTED: uncharacterized protein LOC105642006 isoform X3 [Jatropha curcas] |
| 14 |
Hb_012691_010 |
0.2080170717 |
- |
- |
hypothetical protein POPTR_0005s18750g [Populus trichocarpa] |
| 15 |
Hb_007643_060 |
0.2093923259 |
- |
- |
- |
| 16 |
Hb_002095_050 |
0.2099089853 |
- |
- |
PREDICTED: uncharacterized protein LOC104447479 [Eucalyptus grandis] |
| 17 |
Hb_065133_010 |
0.210886488 |
- |
- |
PREDICTED: receptor-like protein 12 [Vitis vinifera] |
| 18 |
Hb_002942_120 |
0.2118329648 |
transcription factor |
TF Family: bHLH |
Transcription factor ICE1, putative [Ricinus communis] |
| 19 |
Hb_000907_140 |
0.2120590617 |
- |
- |
PREDICTED: annexin D5-like [Jatropha curcas] |
| 20 |
Hb_007943_180 |
0.2127922301 |
- |
- |
PREDICTED: uncharacterized protein LOC105637734 [Jatropha curcas] |