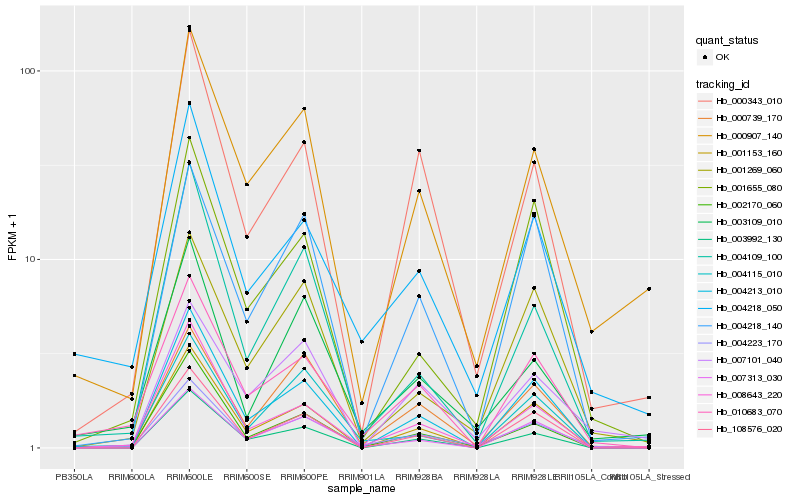
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004223_170 |
0.0 |
- |
- |
PREDICTED: ATP sulfurylase 2 [Jatropha curcas] |
| 2 |
Hb_008643_220 |
0.1329108466 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 [Jatropha curcas] |
| 3 |
Hb_004218_050 |
0.1334872342 |
- |
- |
annexin [Manihot esculenta] |
| 4 |
Hb_002170_060 |
0.1343581086 |
- |
- |
PREDICTED: mitogen-activated protein kinase-binding protein 1 [Jatropha curcas] |
| 5 |
Hb_010683_070 |
0.1344989192 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase CMT2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_007313_030 |
0.1376505222 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 7 |
Hb_001269_060 |
0.1379086762 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000907_140 |
0.1383419259 |
- |
- |
PREDICTED: annexin D5-like [Jatropha curcas] |
| 9 |
Hb_000343_010 |
0.1392007928 |
- |
- |
F3F9.11 [Arabidopsis thaliana] |
| 10 |
Hb_003109_010 |
0.141315025 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 11 |
Hb_000739_170 |
0.1429885335 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 4 [Jatropha curcas] |
| 12 |
Hb_004213_010 |
0.1461300728 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 13 |
Hb_001153_160 |
0.1461640795 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 14 |
Hb_004218_140 |
0.1489467689 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 isoform X3 [Jatropha curcas] |
| 15 |
Hb_108576_020 |
0.1495177567 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003992_130 |
0.1522137949 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_001655_080 |
0.1532263215 |
- |
- |
Purple acid phosphatase precursor, putative [Ricinus communis] |
| 18 |
Hb_004109_100 |
0.1554439109 |
- |
- |
Interactor of constitutive active rops 1 isoform 1 [Theobroma cacao] |
| 19 |
Hb_007101_040 |
0.1559824948 |
- |
- |
PREDICTED: MLP-like protein 423 [Populus euphratica] |
| 20 |
Hb_004115_010 |
0.1560203716 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG11 [Jatropha curcas] |