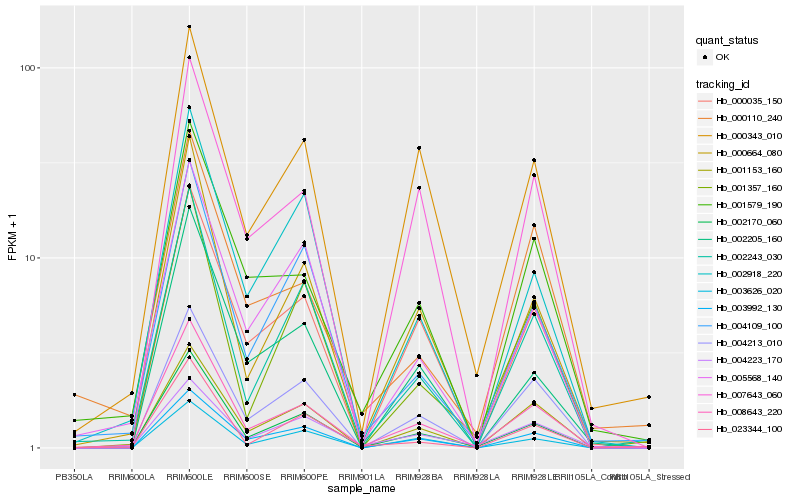
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008643_220 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 [Jatropha curcas] |
| 2 |
Hb_002170_060 |
0.068119721 |
- |
- |
PREDICTED: mitogen-activated protein kinase-binding protein 1 [Jatropha curcas] |
| 3 |
Hb_000664_080 |
0.0874799557 |
- |
- |
PREDICTED: NADP-dependent malic enzyme isoform X2 [Jatropha curcas] |
| 4 |
Hb_001579_190 |
0.103983459 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002243_030 |
0.1116644443 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 3-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000343_010 |
0.1135798988 |
- |
- |
F3F9.11 [Arabidopsis thaliana] |
| 7 |
Hb_003992_130 |
0.1172194411 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_000035_150 |
0.1182939924 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 9 |
Hb_004109_100 |
0.120721019 |
- |
- |
Interactor of constitutive active rops 1 isoform 1 [Theobroma cacao] |
| 10 |
Hb_004213_010 |
0.123162152 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_001153_160 |
0.1240534762 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 12 |
Hb_007643_060 |
0.1311056189 |
- |
- |
- |
| 13 |
Hb_005568_140 |
0.1312279906 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_004223_170 |
0.1329108466 |
- |
- |
PREDICTED: ATP sulfurylase 2 [Jatropha curcas] |
| 15 |
Hb_002918_220 |
0.1347504323 |
- |
- |
PREDICTED: uncharacterized protein LOC105138896 [Populus euphratica] |
| 16 |
Hb_000110_240 |
0.1355566691 |
- |
- |
PREDICTED: uncharacterized protein LOC105642006 isoform X3 [Jatropha curcas] |
| 17 |
Hb_001357_160 |
0.135629689 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002205_160 |
0.1366742589 |
- |
- |
chalcone isomerase [Hibiscus cannabinus] |
| 19 |
Hb_003626_020 |
0.1375033687 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At5g24080 [Jatropha curcas] |
| 20 |
Hb_023344_100 |
0.1377961031 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.2 [Jatropha curcas] |