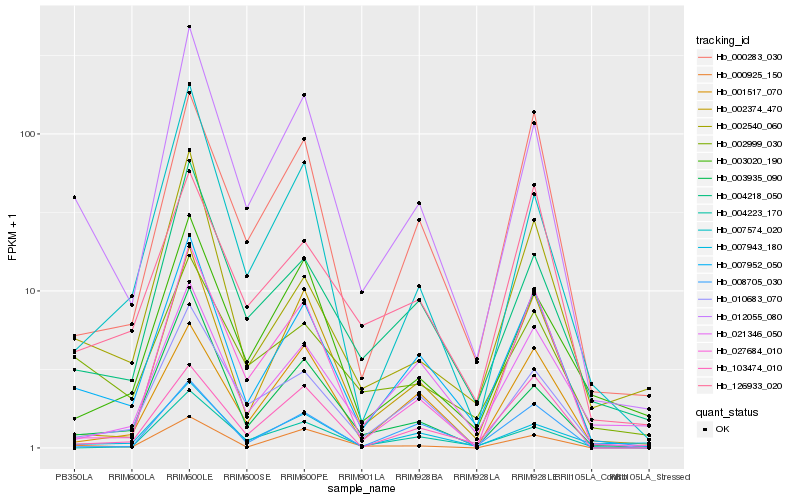
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007952_050 |
0.0 |
- |
- |
PREDICTED: 125 kDa kinesin-related protein-like [Jatropha curcas] |
| 2 |
Hb_012055_080 |
0.1240334677 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 18-like [Jatropha curcas] |
| 3 |
Hb_002374_470 |
0.1377521211 |
- |
- |
PREDICTED: filament-like plant protein isoform X2 [Jatropha curcas] |
| 4 |
Hb_008705_030 |
0.1385534989 |
- |
- |
hypothetical protein JCGZ_13017 [Jatropha curcas] |
| 5 |
Hb_004218_050 |
0.1398325994 |
- |
- |
annexin [Manihot esculenta] |
| 6 |
Hb_010683_070 |
0.1414227259 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase CMT2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000283_030 |
0.1420413419 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA16-like [Populus euphratica] |
| 8 |
Hb_103474_010 |
0.1570592169 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 9 |
Hb_021346_050 |
0.1571770096 |
- |
- |
xylulose kinase, putative [Ricinus communis] |
| 10 |
Hb_007943_180 |
0.1573381709 |
- |
- |
PREDICTED: uncharacterized protein LOC105637734 [Jatropha curcas] |
| 11 |
Hb_001517_070 |
0.1608662943 |
- |
- |
protein with unknown function [Ricinus communis] |
| 12 |
Hb_002540_060 |
0.1651133816 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000925_150 |
0.1655194043 |
- |
- |
PREDICTED: aluminum-activated malate transporter 8-like [Jatropha curcas] |
| 14 |
Hb_126933_020 |
0.1660815412 |
- |
- |
PREDICTED: uncharacterized protein LOC105638525 [Jatropha curcas] |
| 15 |
Hb_004223_170 |
0.1671500987 |
- |
- |
PREDICTED: ATP sulfurylase 2 [Jatropha curcas] |
| 16 |
Hb_027684_010 |
0.1686592515 |
- |
- |
hypothetical protein RCOM_0504250 [Ricinus communis] |
| 17 |
Hb_007574_020 |
0.1696065198 |
- |
- |
PREDICTED: uncharacterized protein LOC105638146 [Jatropha curcas] |
| 18 |
Hb_003935_090 |
0.1722664777 |
- |
- |
PREDICTED: uncharacterized protein At4g38062 [Jatropha curcas] |
| 19 |
Hb_003020_190 |
0.1755091224 |
- |
- |
PREDICTED: methylsterol monooxygenase 2-2 [Jatropha curcas] |
| 20 |
Hb_002999_030 |
0.1762853187 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |