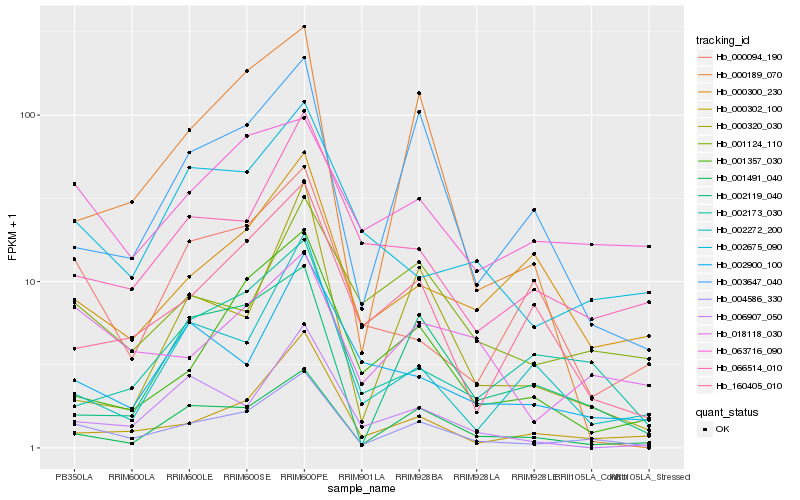
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004586_330 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001491_040 |
0.1696022478 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 3 |
Hb_000302_100 |
0.184841838 |
- |
- |
PREDICTED: uncharacterized protein LOC105766209 [Gossypium raimondii] |
| 4 |
Hb_003647_040 |
0.1929371482 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_018118_030 |
0.2036688206 |
- |
- |
PREDICTED: putative multidrug resistance protein, partial [Musa acuminata subsp. malaccensis] |
| 6 |
Hb_066514_010 |
0.2097623906 |
- |
- |
hypothetical protein RCOM_0204720 [Ricinus communis] |
| 7 |
Hb_002675_090 |
0.2098281193 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_160405_010 |
0.2102471891 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 9 |
Hb_000189_070 |
0.2117743658 |
transcription factor |
TF Family: AUX/IAA |
Auxin-induced protein 22D, putative [Ricinus communis] |
| 10 |
Hb_001357_030 |
0.2117988192 |
- |
- |
hypothetical protein F383_04880 [Gossypium arboreum] |
| 11 |
Hb_006907_050 |
0.2168415584 |
- |
- |
unnamed protein product [Coffea canephora] |
| 12 |
Hb_002119_040 |
0.216872468 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 13 |
Hb_002173_030 |
0.2192491018 |
- |
- |
hypothetical protein JCGZ_12466 [Jatropha curcas] |
| 14 |
Hb_001124_110 |
0.2198788746 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 2-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_000300_230 |
0.2239318625 |
- |
- |
PREDICTED: cation/calcium exchanger 2 [Jatropha curcas] |
| 16 |
Hb_063716_090 |
0.2250002636 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002900_100 |
0.2293191818 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4-like [Jatropha curcas] |
| 18 |
Hb_002272_200 |
0.229985279 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g54790 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000094_190 |
0.2311657389 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 7-like [Jatropha curcas] |
| 20 |
Hb_000320_030 |
0.2325649458 |
- |
- |
PREDICTED: aspartic proteinase-like [Jatropha curcas] |