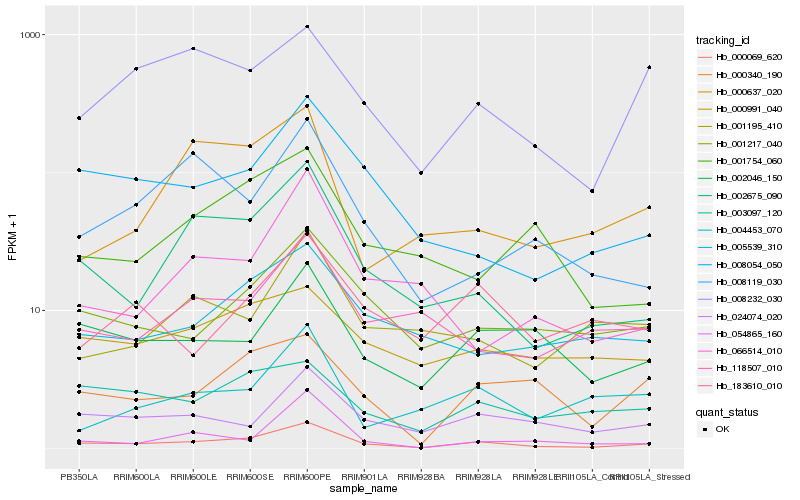
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000069_620 |
0.0 |
- |
- |
serine-threonine protein kinase, putative [Ricinus communis] |
| 2 |
Hb_002675_090 |
0.1673555583 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001217_040 |
0.1825695448 |
- |
- |
hypothetical protein MIMGU_mgv1a0002421mg, partial [Erythranthe guttata] |
| 4 |
Hb_024074_020 |
0.1882554793 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g09600 [Jatropha curcas] |
| 5 |
Hb_002046_150 |
0.1884881983 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g15080 [Jatropha curcas] |
| 6 |
Hb_008054_050 |
0.1890428746 |
- |
- |
PREDICTED: uncharacterized protein LOC101212042 [Cucumis sativus] |
| 7 |
Hb_000340_190 |
0.1920069845 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005539_310 |
0.2033897993 |
- |
- |
basic helix-loop-helix-containing protein, putative [Ricinus communis] |
| 9 |
Hb_008232_030 |
0.2090125851 |
- |
- |
- |
| 10 |
Hb_003097_120 |
0.2099464272 |
- |
- |
PREDICTED: cytokinin dehydrogenase 6 [Jatropha curcas] |
| 11 |
Hb_118507_010 |
0.2171189065 |
- |
- |
hypothetical protein VITISV_019292 [Vitis vinifera] |
| 12 |
Hb_000991_040 |
0.2178534272 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Gossypium raimondii] |
| 13 |
Hb_008119_030 |
0.2200042705 |
- |
- |
PREDICTED: peroxidase 63-like [Jatropha curcas] |
| 14 |
Hb_001195_410 |
0.2204878663 |
- |
- |
PREDICTED: deoxyuridine 5'-triphosphate nucleotidohydrolase [Jatropha curcas] |
| 15 |
Hb_054865_160 |
0.2210394464 |
- |
- |
PREDICTED: D-arabinono-1,4-lactone oxidase-like [Jatropha curcas] |
| 16 |
Hb_001754_060 |
0.2218472605 |
- |
- |
Xyloglucan 6-xylosyltransferase, putative [Ricinus communis] |
| 17 |
Hb_000637_020 |
0.2231855185 |
- |
- |
PREDICTED: uncharacterized protein LOC105650669 [Jatropha curcas] |
| 18 |
Hb_183610_010 |
0.2232455589 |
- |
- |
PREDICTED: uncharacterized protein LOC105635777 [Jatropha curcas] |
| 19 |
Hb_004453_070 |
0.2233875257 |
transcription factor |
TF Family: CPP |
PREDICTED: CRC domain-containing protein TSO1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_066514_010 |
0.2234498356 |
- |
- |
hypothetical protein RCOM_0204720 [Ricinus communis] |