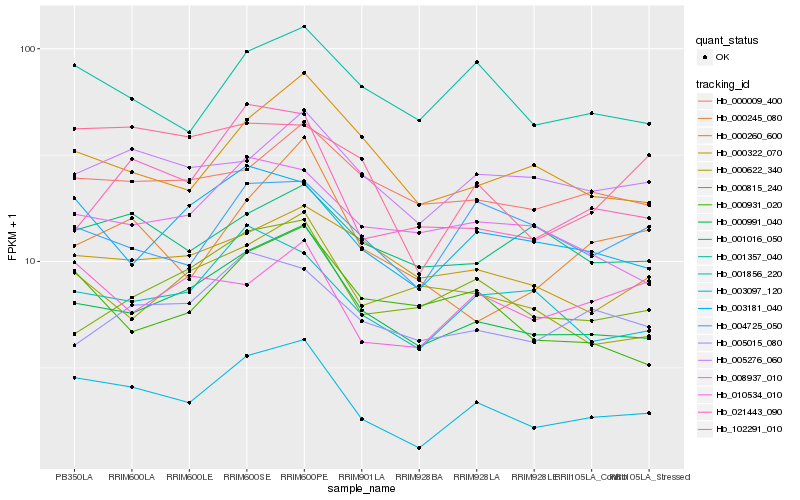
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003097_120 |
0.0 |
- |
- |
PREDICTED: cytokinin dehydrogenase 6 [Jatropha curcas] |
| 2 |
Hb_000991_040 |
0.1075570077 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Gossypium raimondii] |
| 3 |
Hb_003181_040 |
0.137853266 |
- |
- |
PREDICTED: uncharacterized protein LOC105647655 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001856_220 |
0.1389492743 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_102291_010 |
0.1400630898 |
- |
- |
PREDICTED: histone chaperone ASF1B [Populus euphratica] |
| 6 |
Hb_021443_090 |
0.1444101479 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |
| 7 |
Hb_001357_040 |
0.1453518823 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 8 |
Hb_000260_600 |
0.148265843 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_010534_010 |
0.1541202734 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor [Jatropha curcas] |
| 10 |
Hb_005276_060 |
0.1545340658 |
- |
- |
PREDICTED: cyclin-P3-1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000931_020 |
0.1574257908 |
- |
- |
PREDICTED: uncharacterized protein LOC105649774 isoform X1 [Jatropha curcas] |
| 12 |
Hb_008937_010 |
0.1600115962 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80A2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_004725_050 |
0.161679887 |
- |
- |
Rhomboid protein, putative [Ricinus communis] |
| 14 |
Hb_000245_080 |
0.1625063986 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-15 [Jatropha curcas] |
| 15 |
Hb_000815_240 |
0.1636219937 |
- |
- |
PREDICTED: galactoside 2-alpha-L-fucosyltransferase-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_005015_080 |
0.1640496364 |
- |
- |
PREDICTED: shikimate kinase 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000009_400 |
0.1645210252 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g50780 [Jatropha curcas] |
| 18 |
Hb_001016_050 |
0.1647011558 |
- |
- |
drought-inducible protein [Manihot esculenta] |
| 19 |
Hb_000622_340 |
0.1654295124 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase MGP4 [Jatropha curcas] |
| 20 |
Hb_000322_070 |
0.1656342969 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1 [Jatropha curcas] |