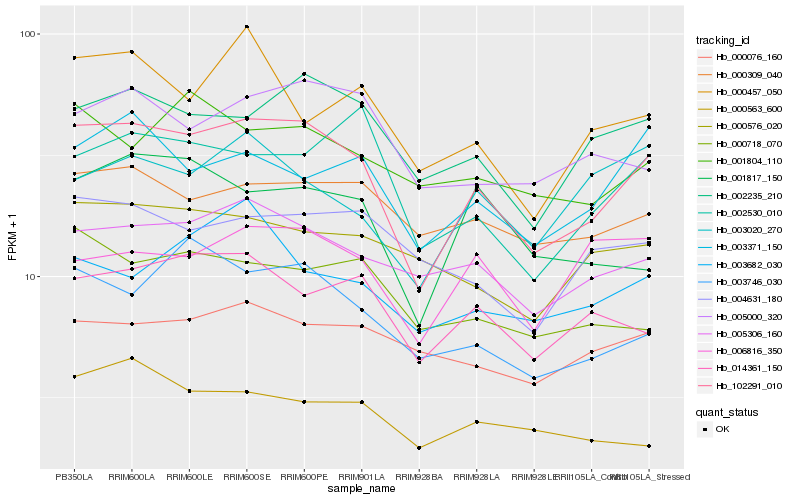
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_102291_010 |
0.0 |
- |
- |
PREDICTED: histone chaperone ASF1B [Populus euphratica] |
| 2 |
Hb_002235_210 |
0.1042056213 |
- |
- |
PREDICTED: vesicle transport v-SNARE 13-like [Populus euphratica] |
| 3 |
Hb_005306_160 |
0.1062777614 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1-like [Jatropha curcas] |
| 4 |
Hb_003746_030 |
0.1111064794 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 2 [Jatropha curcas] |
| 5 |
Hb_014361_150 |
0.1116616111 |
- |
- |
PREDICTED: uncharacterized protein LOC105639827 [Jatropha curcas] |
| 6 |
Hb_003371_150 |
0.1151931074 |
- |
- |
PREDICTED: serine/threonine-protein kinase 10 [Jatropha curcas] |
| 7 |
Hb_000309_040 |
0.1169240427 |
- |
- |
PREDICTED: protein farnesyltransferase subunit beta isoform X1 [Jatropha curcas] |
| 8 |
Hb_005000_320 |
0.1192946053 |
- |
- |
PREDICTED: casein kinase I isoform delta-like [Jatropha curcas] |
| 9 |
Hb_002530_010 |
0.1199314941 |
- |
- |
hypothetical protein EUGRSUZ_C02580 [Eucalyptus grandis] |
| 10 |
Hb_000718_070 |
0.1219032279 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001817_150 |
0.1224472902 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 12 |
Hb_000576_020 |
0.1243119617 |
- |
- |
PREDICTED: uncharacterized protein LOC105638286 [Jatropha curcas] |
| 13 |
Hb_004631_180 |
0.1244027251 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 14 |
Hb_001804_110 |
0.1245723691 |
- |
- |
hypothetical protein CISIN_1g031200mg [Citrus sinensis] |
| 15 |
Hb_000457_050 |
0.1246409691 |
- |
- |
PREDICTED: tether containing UBX domain for GLUT4 [Jatropha curcas] |
| 16 |
Hb_003682_030 |
0.1259979838 |
- |
- |
hypothetical protein CICLE_v10020876mg [Citrus clementina] |
| 17 |
Hb_000076_160 |
0.126936259 |
- |
- |
PREDICTED: AP-5 complex subunit zeta-1 [Jatropha curcas] |
| 18 |
Hb_000563_600 |
0.1270717212 |
- |
- |
PREDICTED: uncharacterized protein LOC104428079 [Eucalyptus grandis] |
| 19 |
Hb_003020_270 |
0.1273308097 |
- |
- |
PREDICTED: ras-related protein Rab2BV [Jatropha curcas] |
| 20 |
Hb_006816_350 |
0.1288720365 |
- |
- |
PREDICTED: syntaxin-43 [Jatropha curcas] |