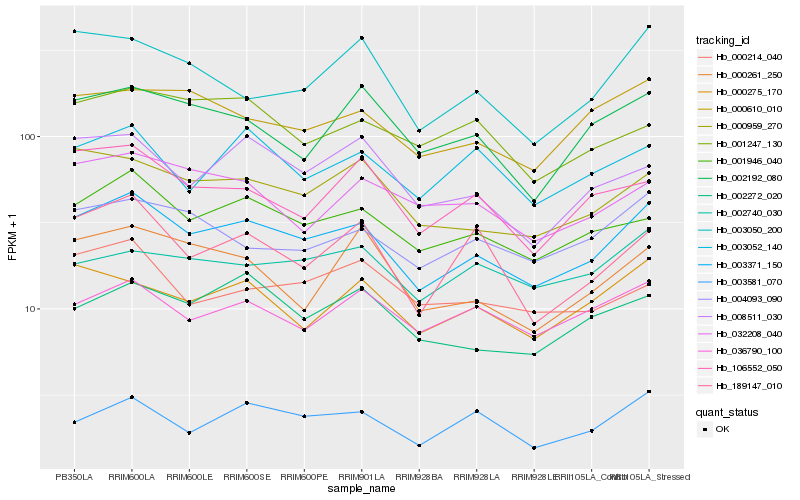
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003371_150 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase 10 [Jatropha curcas] |
| 2 |
Hb_001946_040 |
0.0767567004 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000959_270 |
0.0847949457 |
- |
- |
PREDICTED: uncharacterized protein LOC105641005 [Jatropha curcas] |
| 4 |
Hb_000610_010 |
0.0897097822 |
- |
- |
PREDICTED: SKP1-like protein 1B [Musa acuminata subsp. malaccensis] |
| 5 |
Hb_004093_090 |
0.0918635845 |
- |
- |
PREDICTED: uncharacterized protein LOC105633550 [Jatropha curcas] |
| 6 |
Hb_003581_070 |
0.0923216672 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g40405 [Malus domestica] |
| 7 |
Hb_036790_100 |
0.0931156727 |
- |
- |
PREDICTED: uncharacterized protein LOC105645120 [Jatropha curcas] |
| 8 |
Hb_000275_170 |
0.094591678 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b-like [Jatropha curcas] |
| 9 |
Hb_002272_020 |
0.0960093104 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-3 [Jatropha curcas] |
| 10 |
Hb_008511_030 |
0.097610038 |
- |
- |
guanine nucleotide-binding protein beta, putative [Ricinus communis] |
| 11 |
Hb_106552_050 |
0.0983563999 |
- |
- |
PREDICTED: uncharacterized protein LOC105642134 [Jatropha curcas] |
| 12 |
Hb_002192_080 |
0.0984153018 |
- |
- |
hypothetical protein B456_013G069200 [Gossypium raimondii] |
| 13 |
Hb_002740_030 |
0.0997007893 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003052_140 |
0.1010477806 |
- |
- |
s-adenosylmethionine decarboxylase, putative [Ricinus communis] |
| 15 |
Hb_189147_010 |
0.1017509358 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_032208_040 |
0.1036512599 |
- |
- |
PREDICTED: serine/arginine-rich SC35-like splicing factor SCL33 [Populus euphratica] |
| 17 |
Hb_003050_200 |
0.1041108958 |
- |
- |
Ribosomal protein L18ae/LX family protein [Theobroma cacao] |
| 18 |
Hb_000214_040 |
0.1057749134 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001247_130 |
0.1061887464 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha [Jatropha curcas] |
| 20 |
Hb_000261_250 |
0.106218512 |
- |
- |
elongation factor 1-alpha [Lepidium sativum] |