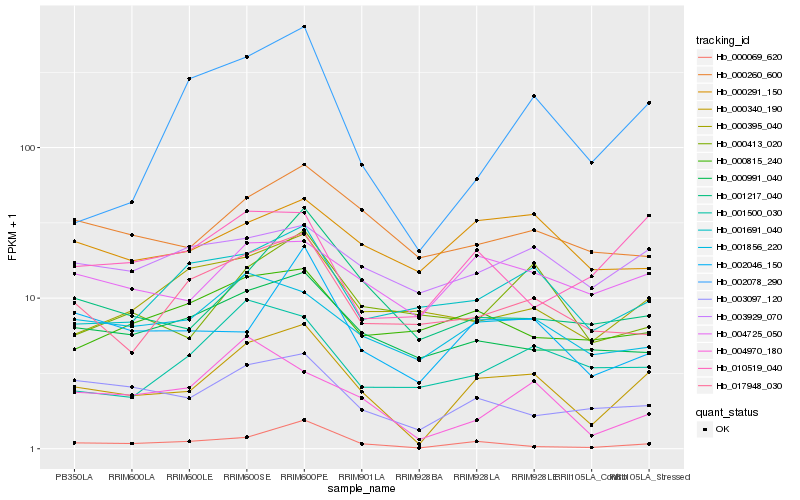
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000340_190 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003097_120 |
0.18746094 |
- |
- |
PREDICTED: cytokinin dehydrogenase 6 [Jatropha curcas] |
| 3 |
Hb_000069_620 |
0.1920069845 |
- |
- |
serine-threonine protein kinase, putative [Ricinus communis] |
| 4 |
Hb_002046_150 |
0.1948635589 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g15080 [Jatropha curcas] |
| 5 |
Hb_004725_050 |
0.1973621608 |
- |
- |
Rhomboid protein, putative [Ricinus communis] |
| 6 |
Hb_001856_220 |
0.1989377003 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000991_040 |
0.2007539813 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Gossypium raimondii] |
| 8 |
Hb_001217_040 |
0.2036045055 |
- |
- |
hypothetical protein MIMGU_mgv1a0002421mg, partial [Erythranthe guttata] |
| 9 |
Hb_001691_040 |
0.2044503706 |
- |
- |
PREDICTED: uncharacterized protein LOC105649546 isoform X2 [Jatropha curcas] |
| 10 |
Hb_017948_030 |
0.2062304818 |
- |
- |
PREDICTED: uncharacterized protein LOC105639493 [Jatropha curcas] |
| 11 |
Hb_000260_600 |
0.2075436627 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000395_040 |
0.2117098678 |
- |
- |
PREDICTED: uncharacterized protein LOC105645576 [Jatropha curcas] |
| 13 |
Hb_003929_070 |
0.2121163914 |
- |
- |
CBL-interacting serine/threonine-protein kinase 1 isoform 3 [Theobroma cacao] |
| 14 |
Hb_004970_180 |
0.2127316405 |
- |
- |
- |
| 15 |
Hb_000413_020 |
0.2137831129 |
- |
- |
PREDICTED: U-box domain-containing protein 8 [Jatropha curcas] |
| 16 |
Hb_001500_030 |
0.2149712526 |
- |
- |
PREDICTED: uncharacterized protein At2g33490 isoform X2 [Jatropha curcas] |
| 17 |
Hb_010519_040 |
0.2177300821 |
- |
- |
PREDICTED: uncharacterized protein LOC105643567 [Jatropha curcas] |
| 18 |
Hb_000291_150 |
0.2184903192 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7 [Jatropha curcas] |
| 19 |
Hb_000815_240 |
0.2218765568 |
- |
- |
PREDICTED: galactoside 2-alpha-L-fucosyltransferase-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_002078_290 |
0.2227284054 |
- |
- |
PREDICTED: uncharacterized protein LOC100852520 [Vitis vinifera] |