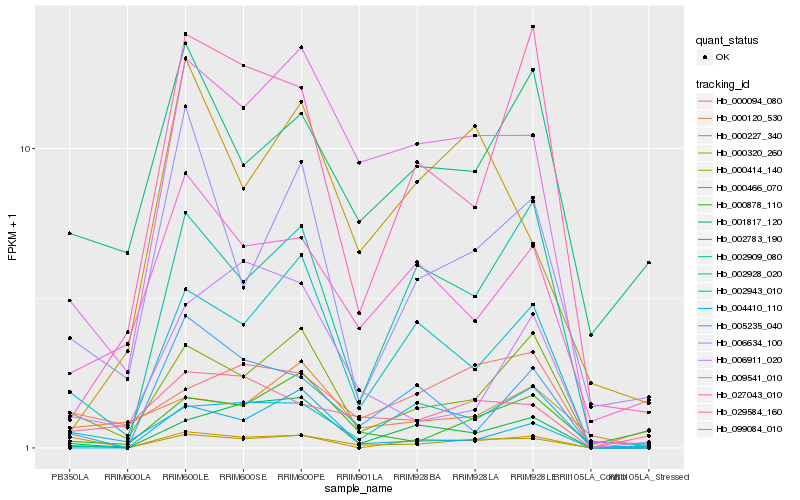
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000414_140 |
0.0 |
- |
- |
Chalcone synthase, putative [Ricinus communis] |
| 2 |
Hb_009541_010 |
0.1387324597 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 6 [Jatropha curcas] |
| 3 |
Hb_000466_070 |
0.1585197868 |
- |
- |
hypothetical protein JCGZ_08989 [Jatropha curcas] |
| 4 |
Hb_002928_020 |
0.1702724076 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 5 |
Hb_001817_120 |
0.1958960691 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 1 [Fragaria vesca subsp. vesca] |
| 6 |
Hb_002909_080 |
0.2126791142 |
- |
- |
inositol or phosphatidylinositol kinase, putative [Ricinus communis] |
| 7 |
Hb_002943_010 |
0.21990183 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 [Nelumbo nucifera] |
| 8 |
Hb_006634_100 |
0.2274092714 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000227_340 |
0.2319760106 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 10 |
Hb_005235_040 |
0.2354855045 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 11 |
Hb_000878_110 |
0.237420316 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 12 |
Hb_029584_160 |
0.237677981 |
- |
- |
PREDICTED: thioredoxin-like 2, chloroplastic [Populus euphratica] |
| 13 |
Hb_000094_080 |
0.2387797796 |
- |
- |
PREDICTED: uncharacterized protein LOC105633792 [Jatropha curcas] |
| 14 |
Hb_027043_010 |
0.2411446046 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase GDPD4 [Jatropha curcas] |
| 15 |
Hb_099084_010 |
0.2421836747 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 16 |
Hb_000320_260 |
0.2433817522 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_004410_110 |
0.2508372497 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_000120_530 |
0.2523458707 |
transcription factor |
TF Family: C3H |
RING finger protein 113A, putative [Ricinus communis] |
| 19 |
Hb_006911_020 |
0.2528260347 |
- |
- |
CC-NBS-LRR protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_002783_190 |
0.253189015 |
- |
- |
PREDICTED: uncharacterized protein LOC105635339 [Jatropha curcas] |