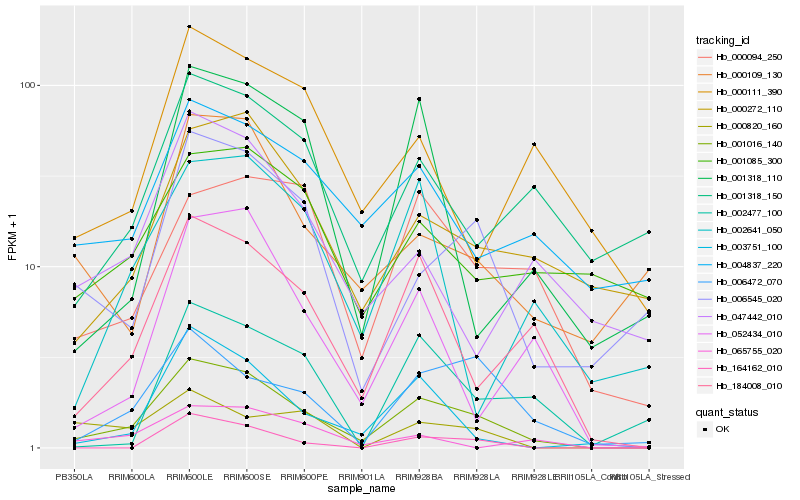
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001016_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647117 [Jatropha curcas] |
| 2 |
Hb_184008_010 |
0.1851450854 |
- |
- |
putative LRR receptor-like serine/threonine-protein kinase [Morus notabilis] |
| 3 |
Hb_003751_100 |
0.2087277246 |
- |
- |
Amino acid permease 2 [Glycine soja] |
| 4 |
Hb_006472_070 |
0.213809436 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 5 |
Hb_006545_020 |
0.2208674975 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 6 |
Hb_000272_110 |
0.2248262278 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 7 |
Hb_052434_010 |
0.229391061 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like serine/threonine-protein kinase At3g14840 [Sesamum indicum] |
| 8 |
Hb_004837_220 |
0.2353347661 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 9 |
Hb_000094_250 |
0.2365452548 |
- |
- |
PREDICTED: uncharacterized protein LOC105633774 isoform X3 [Jatropha curcas] |
| 10 |
Hb_001318_110 |
0.2369018956 |
- |
- |
PREDICTED: uncharacterized protein LOC105642677 [Jatropha curcas] |
| 11 |
Hb_000109_130 |
0.2385150626 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 9 [Jatropha curcas] |
| 12 |
Hb_164162_010 |
0.2449656781 |
- |
- |
PREDICTED: uncharacterized protein At1g08160-like [Jatropha curcas] |
| 13 |
Hb_001318_150 |
0.2457609738 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha-like [Jatropha curcas] |
| 14 |
Hb_002641_050 |
0.2469092302 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 48 [Jatropha curcas] |
| 15 |
Hb_047442_010 |
0.2514693267 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 16 |
Hb_002477_100 |
0.2523759656 |
- |
- |
PREDICTED: protein Brevis radix-like 2 [Jatropha curcas] |
| 17 |
Hb_000111_390 |
0.2542569006 |
- |
- |
PREDICTED: uncharacterized protein LOC105631262 isoform X1 [Jatropha curcas] |
| 18 |
Hb_065755_020 |
0.2558021832 |
- |
- |
choline monooxygenase, putative [Ricinus communis] |
| 19 |
Hb_001085_300 |
0.2569856066 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 20 |
Hb_000820_160 |
0.2573585194 |
- |
- |
- |