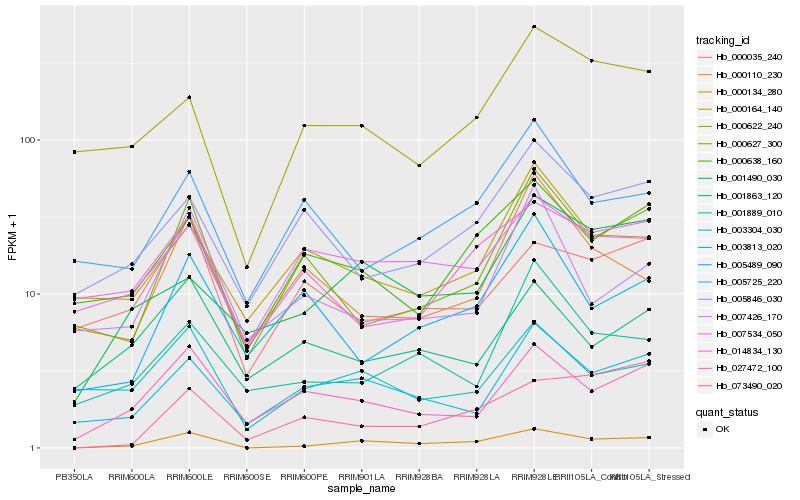
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000134_280 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628677 [Jatropha curcas] |
| 2 |
Hb_003813_020 |
0.2153079506 |
- |
- |
PREDICTED: uncharacterized protein LOC105638736 isoform X1 [Jatropha curcas] |
| 3 |
Hb_027472_100 |
0.2164081082 |
- |
- |
PREDICTED: uncharacterized protein LOC105646173 [Jatropha curcas] |
| 4 |
Hb_000638_160 |
0.2296405005 |
- |
- |
PREDICTED: uncharacterized protein LOC105637288 [Jatropha curcas] |
| 5 |
Hb_001490_030 |
0.2296917153 |
- |
- |
PREDICTED: psbP domain-containing protein 6, chloroplastic [Jatropha curcas] |
| 6 |
Hb_003304_030 |
0.2305565708 |
- |
- |
PREDICTED: uroporphyrinogen-III synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 7 |
Hb_000627_300 |
0.2316707987 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 8 |
Hb_007534_050 |
0.239373368 |
- |
- |
PREDICTED: glutamyl-tRNA reductase-binding protein, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_000622_240 |
0.2406429999 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1A, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001889_010 |
0.2453314418 |
transcription factor |
TF Family: Orphans |
PREDICTED: protein CHLOROPLAST IMPORT APPARATUS 2 [Jatropha curcas] |
| 11 |
Hb_005489_090 |
0.2505941895 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase ICT1, mitochondrial [Jatropha curcas] |
| 12 |
Hb_073490_020 |
0.2509246854 |
- |
- |
hypothetical protein PRUPE_ppa016333mg, partial [Prunus persica] |
| 13 |
Hb_007426_170 |
0.2528990278 |
transcription factor |
TF Family: TCP |
- |
| 14 |
Hb_005846_030 |
0.2534770765 |
- |
- |
PREDICTED: 50S ribosomal protein L29, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001863_120 |
0.2542185741 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000164_140 |
0.2545380866 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 17 |
Hb_005725_220 |
0.2552672792 |
- |
- |
PREDICTED: uncharacterized protein LOC105630808 [Jatropha curcas] |
| 18 |
Hb_000035_240 |
0.2560086837 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000110_230 |
0.256157902 |
- |
- |
Plastid-specific 30S ribosomal protein 3, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_014834_130 |
0.2574858004 |
- |
- |
PREDICTED: formyltetrahydrofolate deformylase 1, mitochondrial-like [Jatropha curcas] |