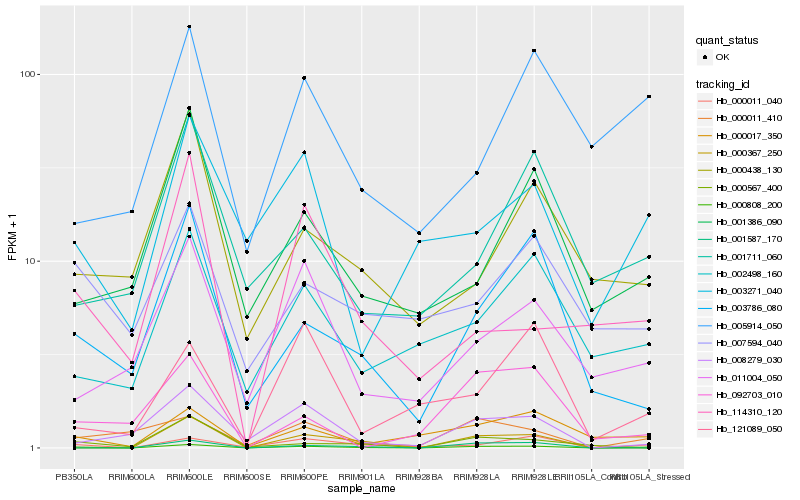
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000808_200 |
0.0 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase CKI1-like [Jatropha curcas] |
| 2 |
Hb_000011_040 |
0.2374319464 |
- |
- |
hypothetical protein JCGZ_24593 [Jatropha curcas] |
| 3 |
Hb_000011_410 |
0.286480651 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000017_350 |
0.2871789515 |
- |
- |
PREDICTED: uncharacterized protein LOC105645799 isoform X2 [Jatropha curcas] |
| 5 |
Hb_003786_080 |
0.2878845479 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 6 |
Hb_008279_030 |
0.2992504058 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000367_250 |
0.3054631382 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_002498_160 |
0.3077719441 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |
| 9 |
Hb_011004_050 |
0.3110485701 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |
| 10 |
Hb_001587_170 |
0.3129669121 |
- |
- |
hypothetical protein B456_010G223200, partial [Gossypium raimondii] |
| 11 |
Hb_003271_040 |
0.3171861426 |
- |
- |
PREDICTED: SEC14 cytosolic factor [Jatropha curcas] |
| 12 |
Hb_092703_010 |
0.3189199243 |
- |
- |
PREDICTED: putative kinase-like protein TMKL1 [Jatropha curcas] |
| 13 |
Hb_001386_090 |
0.3221033977 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |
| 14 |
Hb_000567_400 |
0.3222727246 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 15 |
Hb_005914_050 |
0.3240053767 |
- |
- |
PREDICTED: 50S ribosomal protein L28, chloroplastic [Jatropha curcas] |
| 16 |
Hb_007594_040 |
0.3276110489 |
- |
- |
calcium lipid binding protein, putative [Ricinus communis] |
| 17 |
Hb_121089_050 |
0.3280910738 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000438_130 |
0.3286233982 |
- |
- |
uroporphyrinogen decarboxylase, putative [Ricinus communis] |
| 19 |
Hb_114310_120 |
0.3321230909 |
- |
- |
PREDICTED: 50S ribosomal protein L31, chloroplastic [Eucalyptus grandis] |
| 20 |
Hb_001711_060 |
0.3332199995 |
- |
- |
PREDICTED: ferredoxin isoform X1 [Jatropha curcas] |